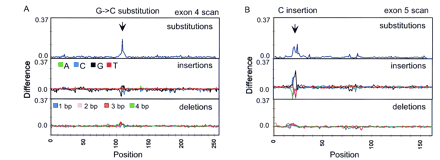
Detection of an insertion in the p53 gene. Eleven exons of the p53 gene were simultaneously scanned (∼1700 bases including primers) byTaq DNA ligation to 8-mer arrays. Two individual samples (11 exons each) were applied to separate arrays with one sample used as a wild-type reference and the other the unknown. Mutation scans searched for substitutions, single-base insertions (A, C, G, T) and multiple-base deletions (1, 2, 3, and 4 bp). (A) A substitution scan (positive envelope) of exon 4 revealed a mutation footprint that correctly identified a G → C base change. The other scans have no appreciable footprints. (B) Mutation scans of exon 5 exhibit footprints in both the substitution (positive envelope) and insertion scans. The insertion scan identified the mutation as a C insertion (increase in the complementary G insertion probes) in the unknown target, whereas the substitution scan provided an ambiguous identification. This insertion was confirmed by conventional sequencing. Normalization of the insertion (A, C, G, T) and deletion probes (1, 2, 3, and 4 bp) was accomplished as for the substitution probes by setting the sum of the four probe intensities to unity.











