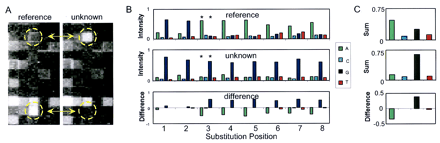
Mutation detection by comparing base-call sets. Two 1.2-kb HIV amplicons differing by eleven single-base substitutions were used as targets. Standard T4 DNA ligation conditions and 8-mer arrays were used for the assay. (A) A small portion of the array images for the reference target (left) and unknown (right) target are shown. The circled probes (interrogating position 435) on each array differ by a single-base substitution at position 3. These probes are indicated by asterisks in B. (B) The normalized intensities of the eight base-call sets interrogating position 435 are shown for both the reference and the unknown target. The intensity differences between these base-call sets are also shown. Note the discordance of the first two base-call sets with the other six sets for the reference target. (C) The composite base-call sets (both strands combined) for the reference and unknown targets and their arithmetic difference (I unknown–I reference) is shown. An A → G substitution at position 435 can be inferred from these data.











