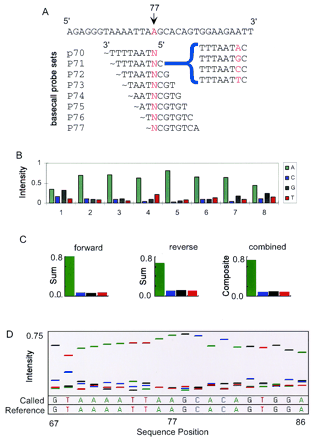
Construction of base-call sets from 8-mer array data by use of a reference sequence. (A) A portion of a 535-bp cftrreference sequence (forward strand) is shown along with the sequences of overlapping probe sets interrogating position 77 within this sequence. A set of four probes differing by a single-base substitution (indicated by red letter N) constitute a base-call set (an example is bracketed). (B) The intensities of the probes within each base-call set are extracted from the array data and plotted. The probes are enumerated such that an A probe actually contains the complementary T base substitution for forward strand interrogations and an A base for reverse strand interrogations. (C) Summation of the interrogation probes (A, C, G, T) across all eight base-call sets yields a summed base-call set for both the forward and reverse strands. A final composite base-call set is derived from these summed sets, from which a final base call is made. (D) A sequence trace is constructed by plotting the normalized intensity values for each composite base-call set as a function of sequence position. Positions 67–86 are plotted for the 535-bp cftr target. The Called bases were generated by our base-calling algorithm using array data, whereas the Reference bases are from the actual sequence of the reference sample.











