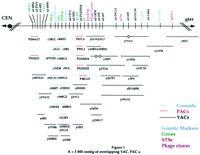
A 3-Mb Contig from D11S987 to MLK3, a Gene-Rich Region in 11q13
Abstract
We have combined genetic, radiation-reduced somatic cell hybrid (RRH), fluorescent in situ hybridization (FISH), and physical mapping methods to generate a contig of overlapping YAC, PAC, and cosmid clones corresponding to >3 continuous Mb in 11q13. A total of 15 STSs [7 genes (GSTP1, ACTN, PC, MLK3, FRA1, SEA, HNP36), 4 polymorphic loci (D11S807, D11S987, GSTP1, D11S913), 3 ESTs (D11S1956E, D11S951E, and WI-12191), and 1 anonymous STS (D11S703)], mapping to three independent RRH segregation groups, identified 26 YAC, 7 PAC, and 16 cosmid clones from the CGM, Roswell Park, CEPH Mark I, and CEPH MegaYAC YAC libraries, a 5 genome equivalent PAC library, and a chromosome 11-specific cosmid library. Thirty-six Alu–PCR products derived from 10 anonymous bacteriophage λ clones, a cosmid containing the polymorphic markerD11S460, or STS-positive YAC or cosmid clones were identified and used to screen selected libraries by hybridization, resulting in the identification of 19 additional clones. The integrity and relative position of a subset of clones was confirmed by FISH and were found to be consistent with the physical and RRH mapping results. The combination of STS and Alu–PCR-based approaches has proven to be successful in attaining contiguous cloned coverage in this very GC-rich region, thereby establishing for the first time the absolute order and distance between the markers: CEN–MLK3–(D11S1956E/D11S951E/WI-12191)–FRA1–D11S460–SEA–HNP36/D11S913–ACTN–PCD11S703–GSTP1–D11S987–TEL.
[On-line supplementary material concerning screening materials and clones referred to in the text as Table 1 is available athttp://genome.wustl.edu/gerhard/gerhard.html or http://www.cshl.org/gr. The sequence data described in this paper have been submitted to the GenBank data library under accession no. AF009361.]
The genetic composition of human chromosome 11q13 includes several disease loci, including those for McArdles’s disease [caused by mutations in muscle glycogen phosphorylase (PYGM; Bartrum et al. 1993)], pyruvate carboxylase deficiency [with mutations in pyruvate carboxylase (PC; Freytag and Collier 1984)], parathyroid adenomatatosis 1 and centrocytic lymphoma [with translocations of cyclin D1 (CCND1; Motokura et al. 1991)], atopy [IgE responsiveness (FCER1B; Young et al. 1992)], and multiple endocrine neoplasia type 1 (MEN1; Chandrasekharappa et al. 1997). Several proto-oncogenes such as fos-related antigen 1 (FRA1; Sinke et al. 1993) and the homolog of the avian S13 virus [sarcoma, erythroblastosis, and anemia (SEA; Williams et al. 1988)] map to this region. In addition to several housekeeping genes, including α-actinin 3 (ACTN; Beggs et al. 1992) and glutathione S-transferase π 1 (GSTP1; Morrow et al. 1989), several growth factors, signal transduction molecules, and proteins potentially involved in the cell cycle, including mixed lineage protein kinase 3 (MLK3; Ing et al. 1994) and a hydrophobic 36-kD protein delayed early response gene (HNP36; Williams and Lanahan 1995; Williams et al. 1997), also map to 11q13. Best’s macular dystrophy (Forsman et al. 1992), Bardet-Biedl syndrome 1 (BBS1; Leppert et al. 1994), insulin-dependent diabetes mellitus 4 (IDDM4; Davies et al. 1994), and spinocerebellar ataxia 5 (SCA5; Ranum et al. 1994) are among the disease loci whose genes reside in this region that yet remain to be identified.
Previously, we have reported the mapping of several new polymorphisms in the 11q13 region (Smith et al. 1995a,b). Here we present a contig of yeast artificial chromosome (YAC), P1 artificial chromosome (PAC), and cosmid clones that extend >3 Mb between D11S987 andMLK3. In the process of identifying overlapping clones, a concerted effort was made to incorporate transcribed sequences, known genes, and genetic markers from the published literature to facilitate the integration of existing resources and information. In doing so, a series of overlapping clones has been created, which makes it possible to correlate physical, radiation hybrid, and genetic distances. This contig will serve as a cloned resource for the isolation of disease loci mapping in this interval.
RESULTS AND DISCUSSION
11q13 is a chromosome band rich in genes that cause disease phenotypes whose causes are still unclear (Shows et al. 1996). The positional cloning approach for the identification of disease genes requires the isolation of the candidate region in recombinant DNA clones. We initiated the cloning of this region from the distal polymorphic locus GSTP1 and ended at the gene MLK3.The Center for Genetics in Medicine (CGM), Centre d’Etude du Polymorphisme Humain (CEPH) Mark I, and chromosome 11-only YAC libraries were screened. Because this region is very under-represented in YAC libraries (see below), other recombinant libraries were added to the screening process as needed.
Contig Construction
We chose two methods for screening: a sequence-tagged site (STS)-based approach (Green and Olson 1990), and an Alu–PCR approach (Liu et al. 1995). PCR-based screens of the CGM (Brownstein et al. 1989) and/or chromosome 11-only (Qin et al. 1993) YAC libraries were conducted using STSs generated from 14 loci (GSTP1, PC, ACTN, HNP36, SEA, FRA1, MLK3, WI-12191, D11S1956E, D11S951E, D11S987, GSTP1, D11S913, and D11S807) reported in the literature, which localize to 11q13 by radiation hybrid (Richard et al. 1991; Larsson et al. 1992; James et al. 1994), genetic linkage (Dib et al. 1996), or tumor amplification mapping (Szepetowski et al. 1992). A select subset of CEPH megabase clones (Dausset et al. 1990) were also screened by STS. A total of 26 YAC clones were isolated (Table 1, located on-line at http://genome.wustl.edu/gerhard/gerhard.html orhttp://www.cshl.org/gr).
The Alu–PCR approach was used in chromosome walks oriented to extend the contig proximal from D11S987. Three sets ofAlu element-specific primers were used in PCR amplification of bacteriophage, YAC, PAC, and cosmid clones. Thirteen 11q13 bacteriophage clones (Gerhard et al. 1992) were used as anonymous DNA segments entry points for clone isolation. Alu–PCR products were used in library hybridization screens of the chromosome 11-only YAC, CEPH Mark I (Albertson et al. 1990) YAC, PAC, and/or cosmid libraries. Under standard PCR conditions, one to eight major inter-Alu–PCR products were identified per YAC clone assayed, facilitating the efficient generation of numerous additional hybridization probes to be used for walking. Thirty-sixAlu–PCR products identified an additional 19 YAC clones. We attained an average STS/Alu–PCR product “landmark” density of 1 marker per 60 kb of DNA, based on the estimate of total distance cloned.
To increase clone depth in the region between PC andACTN, a PAC library (Ioannou et al. 1994) was screened by hybridization and/or PCR for PC and ACTN. We identified four ACTN-positive and one PC-positive PAC clones (Table 1). Fingerprinting analysis of these clones was performed and revealed that P95L2 and P126M24 are completely contained within the two overlapping PACs, P267C6 and P123H18 (data not shown). The chromosome 11 cosmid library (Evans et al. 1989) was screened forFRA1, SEA, HNP36, and D11S460 and D11S913,and corresponding clones were identified for all exceptD11S913.
A contig map representing the best position and sizes of select clones is shown in Figure 1. More than 3 Mb of cloned coverage is estimated based on clone size. Not all of the clones identified for every screen are shown if no new information was contributed with that STS/Alu–PCR. The elimination of some clones because of inconsistent data (potential chimeric or unstable clones) resulted in the generation of a more complete, less complicated map but could have also artificially reduced the size of some of the clones and thereby total estimated coverage.
A >3-Mb contig of overlapping YAC, PAC, and cosmid clones. STS andAlu–PCR product screening reagents are listed above the black line, and positions are identified by vertical lines. Genetic markers are indicated in blue, genes in green, STSs in red, and bacteriophageAlu products in purple. (Phage cluster) Corresponding phage clone-derived Alu–PCR products listed in Table 1. (*ESTs)D11S951E, D11S1956E, and WI-12191. (a1)Alu–PCR products obtained using 263 primers; (a2) PCR products obtained using S/J primers; (a3) PCR products obtained using 278 primers (see Methods). Clones are represented by horizontal lines: red lines represent PAC clones, blue lines represent cosmid clones, and black lines represent YAC clones. Corresponding STS/Alu–PCR content is shown by vertical lines. PAC size is estimated at 125 kb and cosmid size at 40 kb. An open circle identifies a probable internal deletion. Parentheses indicate unstable clones (i.e., strains that have different insert sizes in two separate isolations). To simplify the diagram, not all of the screening reagents/clones used for the establishment of the contig are depicted if the information was redundant. Table 1 lists the screening materials and clones used in the development of the contig depicted here and can be accessed athttp://genome.wustl.edu/gerhard/gerhard.html orhttp://www.cshl.or/gr.
We observed an under-representation of YACs isolated per STS/Alu–PCR products screened versus what is expected, based on estimated haploid genome coverage of the libraries screened. In addition, an average YAC clone size of 230 kb of the clones isolated is smaller than the average clone size of the library. These trends may reflect inherent genomic instability of sequences within this region, which has been reported in clones originating from distal 11q13 (Szepetowski et al. 1996).
Fluorescent In Situ Hybridization
Two-color fluorescent in situ hybridization (FISH) was used to confirm the order of clones in the contig. Biotin- or digoxigenin-labeled Alu–PCR products from y14F8, y19G11, y14D6, y776.e2, y6B2, and yB119H4 were hybridized to metaphase spreads. The clones yB119H4 and y14F8 were used as proximal and distal anchors, respectively. YAC clone yB119H4 resides within the PYGMsegregation group, whereas y14F8 contains the fibroblast growth factor 3 (FGF3) STS and maps to the FGF3 segregation group distal to GSTP1 by RRH. All of the YAC clones map uniquely to chromosome 11q13. Fractional length measurements (Flpter) expressed as the ratio of the distance between the short arm terminus and the signal to the total length of the chromosome were calculated for each probe. The following order was established: cen–[yB119H4–y6B2]–[y14D6–y776.e2]–y19G11—y14F8—qter. The order of the YACs within the brackets could not be determined. The FISH-established order is consistent with the order generated by mapping experiments.
Radiation-Reduced Somatic Cell Hybrids
All markers were mapped by radiation-reduced somatic cell hybrids (RRHs) to segregation groups assigned by previously mapped loci (Gerhard et al. 1992; Smith et al. 1995a). MLK3 cosegregates with PYGM; FRA1, D11S913, HNP36, D11S460, PC, ACTN, D11S703, D11S951E, D11S1956E, and WI-12191 with SEA; andD11S987 and D11S807 (Alu–PCR) withGSTP1. Genetic markers (PYGM, D11S913, D11S460, D11S987, and D11S807) used previously in family studies (Nakamura et al. 1989; Larsson et al. 1992; Thakker et al. 1993;Leppert et al. 1994; Weber et al. 1994; Smith et al. 1995b), as well as genes/expressed sequence tags (ESTs) (MLK3, FRA1, PC, ACTN, D11S951E, D11S1956E, WI-12191, HNP36, SEA, and GSTP1) were included to integrate the transcription, genetic, and RRH maps.
An Alu–PCR product from the D11S807 cosmid cCLGW39 maps to the GSTP1 segregation group. However, theD11S807 STS isolated from the end of the D11S807containing cosmid cCLGW39 also maps to J1-11, a short arm containing somatic cell hybrids, as well as to the RRH cell hybrids 3A1, 7C1, 2C2, and 33B1. This result is in disagreement with the previously reported placement by pulsed field gel electrophoresis (PFGE), which suggested that D11S807 is within 900 kb of D11S427 (Weber et al. 1994). We place D11S427 within the PYGMsegregation group by RRH (D.S. Gerhard and C.M. Smith, unpubl.). Library screens with an Alu product from D11S807 did not identify any clones within the contig extending proximal fromD11S987 to MLK3. Therefore, the location ofD11S807 does not appear to map to the region betweenD11S987 and MLK3 and is outside of this contig. Our data suggest a more distal location, within the GSTP1segregation group, distal to the D11S987 locus as the most likely position. Similar examples of sequences that map to both the long and short arm of chromosome 11 have been reported (Qin et al. 1996), suggesting that the D11S807 cosmid may represent a chromosome 11-specific duplication or be chimeric.
Contig Size
The size of the contiguous cloned region extending fromD11S987 to MLK3 based on clone size is >3 Mb. This is more than twice what was estimated by PFGE (Janson et al. 1991;Tanigami et al. 1992; Weber et al. 1994). However, the genetic distance between PYGM and GSTP is 5 cM (Smith et al. 1995a), whereas RRH mapping suggests that SEA and GSTP are separated by at least 1.6 Mb (James et al. 1994). These data support our length estimate.
In conclusion, a series of overlapping YAC, PAC, and cosmid clones spanning at least 3 Mb of 11q13 has been developed. The STS andAlu–PCR methods of library screening were used coordinately to identify clones, develop walking resources, and establish physical linkage between clones. A total of 45 YAC, 7 PAC, and 16 cosmid clones representing 14 STSs and 36 Alu–PCR products were isolated from a number of recombinant libraries resulting in at least a two-clone depth of coverage for all loci but two (Fig. 1;a2-17C1 and D11S987). A combination of RRH, genetic linkage, and FISH mapping was used to provide an independent confirmation of the results. The establishment of this contig containing genes and ESTs incorporates a transcription map within the physical map and allows for the resolution of a number of marker order inconsistencies found in the literature (Larsson et al. 1992; Weber et al. 1994). The average density of one probe per 60 kb along the cloned resource provides an excellent framework map for the initiation of large-scale genome sequencing efforts in this very gene-rich region and the precise mapping of additional ESTs made available by the IMAGE consortium.
The development of this contig has created the resource required for the integration of meiotic mapping information available of use for disease gene mapping. A composite map integrating information obtained from genetic mapping, contig assembly, FISH, and RRH mapping establishes the order CEN–D11S913–D11S460–GSTP1–D11S987–D11S97–FGF3 for the important genetic markers in the region.
Finally, the development of this contig within 11q13 is of utility for the elucidation of disease loci that localize to 11q13, including those for BBS1, IDDM4, and SCA5.
MATERIALS AND METHODS
YAC Libraries
The following lists the YAC resources used, haploid genome coverage, and average clone size, estimated by the original authors: 6-hit CGM, (350 kb; Brownstein et al. 1989); 7-hit CEPH Mark I (430 kb;Albertson et al. 1990); 10-hit CEPH MB (900 kb; Dausset et al. 1990); 3-hit chromosome 11-only (325 kb; Qin et al. 1993).
Preparation of YAC Libraries for Screening
The CGM YAC libraries were prepared for STS screening as described (Imai and Olson 1990). The CEPH Mark I library was prepared for Alu–PCR screening as described (Moir et al. 1993; Liu et al. 1995). A copy of the chromosome 11-only YAC library was pooled for use as template for STS screens by PCR or for the generation ofAlu–PCR products for use in filter hybridization (details from authors upon request). Select clones from the CEPH Megabase library were screened by either method.
Cosmid and PAC Libraries
A copy of a chromosome 11-specific cosmid library (Evans et al. 1989) represented by 17,000 clones or 5 genome equivalent haploid genome coverage was stamped on nylon filters and screened by hybridization.
Gridded high-density hybridization filters representing 15,000 PAC clones of average insert size 130–150 kb were purchased from P. de Jong (Ioannou et al. 1994).
Sources of STSs and Alu–PCR Products
Chromosome 11-specific Bacteriophage Clones
Thirteen bacteriophage clones from a J1-9 chromosome 11-specific bacteriophage λ library, which map to two RRH segregation groups, were used as templates for Alu–STS probe generation (Gerhard et al. 1992).
Alu–PCR Products
Primers S,J, 263-283.1, 263-283.2 are as described (Liu et al. 1995) and the 278 primer is as described (Nelson et al. 1989).Alu–PCR products are designated as “a1” if they were generated using the 263-282.1/2 primers; “a2” if generated with S/J, and “a3” if primed with 278 primers. The PCR was done on 50 ng of template DNA in standard Perkin Elmer buffer for 35 cycles of 92°C for 30 sec, 63°C for 1 min, and 72°C for 2 min.
STSs
STS’s were developed for the following genes/loci: MLK3(277 bp; Ing et al. 1994); WI-12191 (150 bp; WI/MIT Genome Center); D11S1956E (136 bp) and D11S951E (227 bp;Polymeropoulos et al. 1992); FRA1 (102 bp; Sinke et al. 1993);SEA (130 bp; Williams et al. 1988); HNP36 (1.1 kb;Williams et al. 1997); D11S913 (221–227 bp) andD11S987 (82–116 bp; Weissenbach et al. 1992); α-ACTN (127 bp; Beggs et al. 1992); D11S703 (166 bp; Miwa et al. 1993); PC (302 bp; Freytag and Collier 1984);GSTP1 (509 bp; Morrow et al. 1989); D11S807(F-GTTCAGTGATGTAACCATAC, R-ATTCTTTTTCCCTTTCCCAG; 114 bp; S. Zhu, pers. comm.). The GenBank accession number is AF009361.
cDNA Probes
cDNAs for GSTP1 (Morrow et al. 1989), HNP36(Williams et al. 1997), SEA (Williams et al. 1988; Szepetowski et al. 1992; P. Gaudray, pers. comm.), and MLK3 (Washington University–Merck EST Sequencing Project) were used as hybridization probes. D11S807 (cCLGW39) was provided by C. Larsson (Larsson et al. 1992).
Hybridization of Library Filters
STS-specific PCR products, cDNAs, and Alu–PCR probes were radiolabeled with [α-32P]dCTP by random priming (Feinberg and Vogelstein et al. 1983). Alu–S/J probes were preassociated with sheared human placental DNA. Hybridization and washing were carried out as described (Gerhard et al. 1992).
RRHs
Chromosome 11 somatic cell hybrid and RRH lines characterized previously (Gerhard et al. 1992; Smith et al. 1995a) were used for segregation analysis of STSs, Alu–PCR, and end-clone fragments.
Clone End Isolation
Vector–insert junctions were isolated from a select subset of YAC, PAC, and cosmid clones by a modification of vectorette PCR described (Riley et al. 1990) or by Alu–vector PCR (Nelson et al. 1989; D.S. Gerhard and C.M. Smith, unpubl.). The isolated end fragments were used as hybridization probes on a chromosome 11 RRH panel of EcoRI-digested DNAs to identify chimeras.
SEGMAP
Library screen, STS, and clone data were managed using Hypercard version 2.1. Contigs were assembled using the computer analysis program SEGMAP Version 3.20 (C. Magness, Y. Hu, and P. Green, unpubl.). The SEGMAP output integrates the data to formulate a best fit, which is represented schematically to detail screen information, clone size, overlaps between clones, potential chimeric clones, and potential problems including inconsistent or ambiguous data. Potentially problematic clones were eliminated from further analysis when sufficient and consistent clone depth had already been established. In most instances, this meant two to three clones from at least two different libraries were identified for that particular STS orAlu–PCR product.
Fingerprinting
Fingerprinting was performed on the PAC clones according to Washington University Genome Sequencing Center Protocols.
FISH
FISH probes were prepared from YACs as Alu–PCR amplification products using a pool of S/J, 263-282.1/2, 278, andAlu1 + Alu2 (D. Nelson, pers. comm.) products. Amplification products from individual PCRs were combined and precipitated. The YAC-derived Alu–PCR pools were biotin 11–dUTP (Boehringer Mannheim) and digoxigenin 11–dUTP (Boehringer Mannheim) labeled in a 50-μl reaction by nick translation with DNaseI/DNA polymerase I enzyme mix (GIBCO BRL).
Metaphase spreads were processed from thymidine/BrdU-synchronized cell cultures of mitogen-treated peripheral blood from normal individuals as described (Lichter et al. 1988). Interphase two- and three-color FISH was performed using methodologies as described (Trask et al. 1991). To confirm the localization and to permit comparison of the direct R-banded FISH mapping data with previous mapping data obtained by contour length measurements (Lichter et al. 1990) fractional length measurements were performed.
Acknowledgments
We thank Drs. D. Schlessinger, B. Brownstein, V. Stanton, and D. Housman, for providing access/copies of YAC libraries; Dr. L. Deaven for providing a copy of the cosmid library; Dr. J. Williams for clones and unpublished sequence, and Dr. C. Larsson for cosmid clones. We acknowledge the technical support of B. Muneer, P. Taillon-Miller, the help of Drs. T. Kamarasova and M. Marra, and thank Dmitry Reznikov for placing the supporting material on our web site. This work was supported by American Cancer Society grant CN-64C and Retinitis Pigmentosa Foundation grant to D.S.G., and National Institutes of Health training grant 5-P32-HG0002 to C.M.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL gerhard{at}genetics.wustl.edu; FAX (314) 362-7855.
-
- Received February 13, 1997.
- Accepted June 10, 1997.
- Cold Spring Harbor Laboratory Press