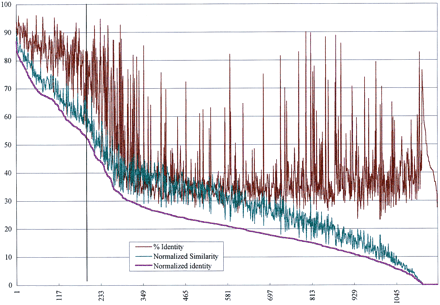
Summary of alignments from similarity searches between 1157 M. leprae proteins (including all of the gene products from this study) and 1564 M. tuberculosis proteins from GenPept. Each of the M. leprae proteins was searched against the set of M. tuberculosis proteins using an implementation of the Smith–Waterman algorithm with default parameters on a Biocellerator (Compugen) in conjunction with the GCG Wisconsin Package. The Normalized Similarity and % Identity values were obtained from the best alignment for each M. leprae protein by multiplying by the fraction of query amino acids represented in each alignment (no. of query residues in alignment/total query length). This was done to provide a better indication of the overall similarity of each M. leprae protein to the best M. tuberculosis homolog. The resulting values were termed Normalized Identity and Normalized Similarity. The pairs were sorted according to the Normalized Identity values in descending order, and the normalized values were plotted together with the raw percent identity values (for comparison) on a graph.











