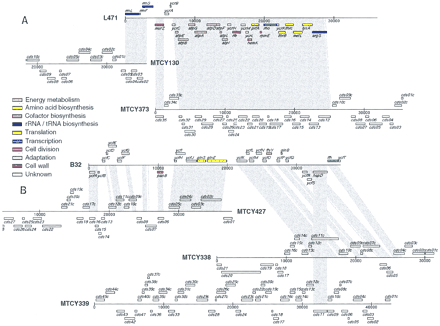
DNA-level matches between two M. leprae cosmids from this study and M. tuberculosis cosmids (Sanger Center, GenBank). The alignments show two particular examples from an exhaustive comparison of the set of M. leprae cosmids reported here against all available M. tuberculosis cosmids (see text). The shading indicates regions of significant similarity between each pair of cosmids. The alignments for cosmid L471 spanned a total of 26,302 nucleotides with 75%–94% identity; those for cosmid B32 spanned 24,009 nucleotides with 71%–85% identity. The M. lepraeORFs are color coded according to function as indicated. The sequences were compared and aligned using Cross_match, an implementation of the Smith–Waterman algorithm developed by P. Green (University of Washington, Seattle). Alignments with >60% identity were sorted using Matchtable (P. Richterich, unpubl.) and examined using a Web browser. A table summarizing the positions of aligned segments between each pair of cosmids was assembled; it was read by a Perl-tk script, Cosmid_map (R. Gibson, unpubl.) in conjunction with two other tables similar to Table 1 (but sorted by cosmid) summarizing the positions of coding frames and functional information (if available) for the M. leprae and M. tuberculosis cosmids.











