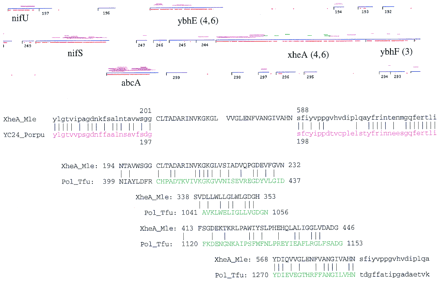
Analysis of the xheA intein. The tightly coupled operon shown is right-to-left 5′ to 3′: ybhF, xheA, ybhE, abcA, nifS, nifU shown in GenomeBrowser format. ORFs longer than 50 codons (blue horizontal lines) have stop codons indicated by short vertical black lines. Magenta horizontal lines above each ORF indicate matches to the NR database (Altschul et al. 1990) with significant BLASTP scores (P < 0.001), where the vertical displacement indicates the percent amino acid identity for that sequence segment. The red lines below the ORFs indicate quality of dicodon usage. Frame number, accession numbers, and gene names based on sequence similarity are in the text below the red lines. The xheA gene is located in M. leprae cosmid B1496 from nucleotide position 2020 to 9152. The amino- and carboxy-terminal regions have strong matches with eukaryotic, prokaryotic, and archaebacterial URFs (unknown function readingframes), including sp | P51240 | YC24_PORPU, and gi | 1742763 (E. coli) at 30%–42% identity (P < 1E-22) as does the central intein region (where intein BLASTP segments are in green to contrast with the normal magenta). The paralogous (intragenomic M. leprae) xheA–ybhF (gi | 466874) duplication is 24% identity, P = 2E-15. The numerals in parentheses represent the ORF numbers for a related cyanobacterial gene cluster (D64004). The sequence alignments (below) indicate the shift in amino acid identity pattern and the conserved motifs at the intein boundaries and internally.











