Single-minded—Two Genes, Three Chromosomes
Comparative sequence and expression analyses of homologous genes from different species allow us to infer their conserved or divergent gene function. This is highlighted by the comparison of thesingle-minded (sim) genes, which have been identified in Drosophila (Crews et al. 1988; Thomas et al. 1988), mouse (Ema et al. 1996; Fan et al. 1996; Moffett et al. 1996; Yamaki et al. 1996), and now in human (Chrast et al., this issue). Decipheringsim gene function is of particular interest because of its importance in the development of the central nervous system (CNS), and the potential involvement of one of the two human SIM genes in the pathogenesis of Down syndrome (DS).
DS is the most frequent genetic cause of mental retardation. Although most cases of DS are attributable to the presence of three full copies of chromosome 21, rare DS patients carry chromosomal rearrangements resulting in triplication of only part of chromosome 21. Molecular characterization of these “partial trisomy” cases has allowed the delineation of a DS critical region (DSCR), located at the sub-band 21q22.2, which correlates with many DS abnormalities (Delabar et al. 1993). Several laboratories have characterized and cloned the DSCR. Using the exon-trapping technique to isolate potential coding sequences within this region, two groups have identified exons that predict an open reading frame that is highly homologous to the Drosophila sim gene product (Chen et al. 1995; Dahmane et al. 1995). In parallel, two murine homologs of sim have been cloned and named Sim1 and Sim2 (Ema et al. 1996; Fan et al. 1996; Moffett et al. 1996; Yamaki et al. 1996). Chrast et al. (this issue) now report the cloning of human SIM1 and SIM2cDNAs. Based on its chromosomal location and sequence, the gene that maps to the DSCR is the human equivalent of the murine Sim2gene (Muenke et al. 1995).
In Drosophila, sim is expressed in the CNS midline cells. These cells, which consist of both glia and neurons, fail to develop insim mutant flies (Crews et al. 1988; Thomas et al. 1988). In contrast, when ectopically expressed, sim can convert the lateral CNS cells to become the CNS midline cells (Nambu et al. 1990). These observations have led Steve Crews and his colleagues to conclude that sim functions as a “master regulator” for theDrosophila CNS midline cell development.
Consistent with its function, sim belongs to the basic helix–loop–helix (bHLH) family of transcription factors, which are known to control the differentiation of various cell types (for review, see Jan and Jan 1993). Immediately carboxy-terminal to its bHLH domain, Sim possesses another conserved domain, termed PAS, an ∼250 amino acid sequence originally found in Period,ARNT, and Sim proteins (Hoffman et al. 1991). Since then, many other bHLH–PAS proteins have been identified. Molecular dissection of bHLH–PAS proteins has led to four primary conclusions (for review, see Hankinson 1995; Schmidt and Bradfield 1996): (1) The basic domain contributes to sequence-specific DNA binding; (2) the HLH and the PAS domains together form a dimerization interface; (3) ARNT seems to be the universal dimerization partner for other bHLH–PAS proteins; and (4) amino acid sequences carboxy-terminal to each bHLH–PAS domain appear to participate in transcriptional activation or repression of the target genes.
Murine and fly SIMs can dimerize with ARNT (Swanson et al. 1995; Ema et al. 1996; Probst et al. 1997) and bind to their cognitive DNA sequences, called the CME (CNS midlineelement; Wharton et al. 1994). However, although the fly Sim activates transcription through its carboxyl terminus (Franks and Crews 1994), the murine SIMs function as repressors in transient transfection assays (Ema et al. 1996; Probst et al. 1997; P. Moffet and J. Pelletier, pers. comm.). Consistent with their divergent activities, the carboxyl termini of fly and murine SIMs are not conserved. Interestingly, as shown by Chrast et al. (this issue), the humanSIM genes are highly homologous to their mouse counterparts throughout the entire coding region, suggesting that they not only also dimerize with ARNT but also act as repressors. It remains to be determined, however, whether the SIM proteins function exclusively as repressors in mammals.
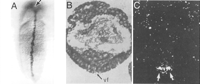
Despite their seemingly different transcriptional roles, the expression patterns of murine Sim genes during development are reminiscent of those of the Drosophila sim (Dahmane 1995; Ema et al. 1996; Fan et al. 1996; Yamaki et al. 1996). For example, Figure1 shows that the Drosophila sim transcript and protein are detected in the CNS midline cells located in the ventral body wall. In conjunction with this, the mouseSim2 is expressed in the developing ventral diencephalon, including its midline. In contrast, Sim1 is expressed in regions immediately adjacent to the ventral midline of the diencephalon and of the spinal cord. Loss- and gain-of-function experiments will be required to address whether the murine homologs also direct the development of cells located adjacent to or within the CNS midline. Interestingly, preliminary results indicate that Sim1 mutant mice generated by homologous recombination display neurological defects (T. Rosenquist, J. Michaud, G. Martin, M. Tessier-Lavigne, and C.-M. Fan, unpubl.). Although mice carrying a Sim2 mutant allele have been generated, their phenotype has not yet been determined (M. Shamblott, A. Lawer, J. Michaud, J.D. Gearhart, and C.-M. Fan, unpubl.).
Comparison of the expression patterns of the sim genes. (A) Drosophila sim protein is expressed in the cells at the ventral midline (indicated by the arrow). (B,C) Bright-field and dark-field views, respectively, of a transverse section of a Drosophila embryo of the same stage as that inA. The sim transcripts are found in the midline cells (white arrows) at both sides of the ventral furrow (vf). (D,E) Transverse sections of an embryonic day (E) 9.5 mouse brain at the level of the diencephalon (di). The mouse Sim2 is expressed in the ventral diencephalon including its midline (D), whereasSim1 is expressed in cells adjacent to the midline (E). (F) Transverse section of an E10.5 mouse spinal cord shows that Sim1 is expressed in cells at both sides of the floor plate (fp), the ventral midline of the spinal cord. [A–C are adapted, with permission, from Thomas et al. (1988).]
The fact that SIM2 maps to the DSCR raises the possibility that it contributes to the DS phenotype when triplicated. Mouse models have been generated by a number of groups to gain insight into the molecular basis of DS. In particular, the Ts65Dn mouse carries a reciprocal translocation, which results in the triplication of a segment of chromosome 16 syntenic to the DSCR and containing Sim2 (Reeves et al. 1995). Neurological abnormalities have been reported in this mouse, including abnormal behavior (Reeves et al. 1995) and age-related degeneration of some cholinergic neurons, a phenotype also observed in DS brains (Holtzman et al. 1996). The use of YACs (yeastartificial chromosomes) in mouse transgenesis will be an elegant approach to test whether dosage imbalance of Sim2 contributes to the Ts65Dn mice phenotype, as it has the advantages of approximating the gene dosage found in DS and in Ts65Dn while assuring appropriate spatiotemporal expression of the transgene (Lamb et al. 1993; Smith et al. 1997).
One fascinating aspect of the Sim genes is that their activities may be further regulated by other means. For example, one of the best characterized bHLH–PAS proteins, the aryl hydrocarbon receptor (AHR), is an inducible transcription activator when bound to the ligand dioxin (for review, see Hankinson 1995; Schmidt and Bradfield 1996). AHR transduces dioxin signal in a manner similar to that of the glucocorticoid receptor (GR): Both of them associate with the molecular chaperone HSP90 and localize in the cytoplasm. Upon ligand binding, both receptor–ligand pairs translocate into the nucleus to activate specific sets of target genes—except that in the nucleus, AHR/dioxin would heterodimerize with ARNT for its DNA binding/transcriptional activity whereas GR homodimerizes. Although AHR is the only PAS protein known to interact with a specific ligand, it is possible that transcriptional activities of the SIM proteins are also regulated by a specific ligand(s). That SIM proteins can also associate with HSP90 supports this hypothesis (McGuire et al. 1995; Probst et al. 1997). In addition to AHR, other PAS domain proteins are also involved in controlling physiological processes in response to the environment, such as circadian rhythm, including the recently identified CLOCK protein in mice (Yu et al. 1987; Antoch et al. 1997; Crosthwaite et al. 1997; King et al. 1997), hypoxic response (Wang et al. 1995; Tian et al. 1996; Ema et al. 1997; Hogenesch et al. 1997), and sporulation (Perego et al. 1989). It is tempting to speculate that these responses are also mediated through specific ligands generated during the day/night transition, hypoxic condition, and nutrient deprivation.
Not only do aryl hydrocarbon and steroid receptors use similar signaling mechanisms, but they can also interact with each other. For example, AHR and the estrogen receptor interfere with each other’s activity (Kharat and Saatcioglu 1996). Moreover, the bHLH–PAS proteins SRC-1 (Onãte et al. 1995) and GRIP-1/TIF-2 (Voegel et al. 1996;Hong et al. 1997) have been shown to associate with and function as coactivators for the steroid receptors in response to hormone binding. These observations raise the intriguing possibility that the SIM proteins may interact with members of the steroid receptor family to coordinate gene expression in a previously unsuspected manner.