A Differential Hybridization Scheme to Identify Photoreceptor-Specific Genes
- Deborah A. Swanson1,
- Carol L. Freund1,
- Jason M. Steel2,
- Shunbin Xu1,
- Lynda Ploder3,4,
- Roderick R. McInnes3,4, and
- David Valle2,5
- 1Predoctoral Training Program in Human Genetics and 2Howard Hughes Medical Institute, Department of Pediatrics, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, and 3Program in Developmental Biology and 4Department of Genetics, Hospital for Sick Children, Toronto, Ontario, Canada M5G 1X8
Abstract
Identification of genes expressed preferentially or exclusively in photoreceptors will facilitate the understanding of photoreceptor biology as well as provide candidate genes for inherited retinal degenerations. To achieve this goal we performed a differential hybridization screen of 3717 well-isolated phage clones from a human retinal cDNA library. Clones were selected for further study if they hybridized exclusively or strongly preferentially to a probe derived from RNA isolated from the cone-predominant retina of 13-line ground squirrels as compared to a probe derived from human fibroblast RNA. Twenty percent of clones (9/45) identified by this screen were derived from photoreceptor-specific genes and an additional 24.4% (11/45) were from neural-specific genes, demonstrating the utility of this strategy in identifying genes important for retinal biology.
[The sequence tags of cDNAs identified in this screen have been deposited in GenBank under accession nos. U89715, U89878–U89888, andU89951–U89955.]
The retinal photoreceptor is an exquisitely specialized cell. In response to a single photon of light, it produces an electrochemical signal that is transmitted, via other cells of the neural retina, to the brain, where the signal is integrated with others and perceived as vision. Photoreceptors are of two major types, rods and cones (95% and 5% of the total in humans, respectively). Rods mediate dim-light vision while cones are responsible for vision in bright light as well as color vision and high acuity vision (Records 1979; Dowling 1987; Hubel 1988). Establishment and maintenance of photoreceptor identity and function depend on the expression of a unique set of genes. Identification of these photoreceptor-specific genes should facilitate the understanding of photoreceptor biology as well as provide candidate genes for inherited retinal degenerations.
A direct way to clone genes expressed specifically in retina is to use information derived from known retinal-specific proteins. For example,Nathans and Hogness (1983) utilized amino acid sequence information from bovine rhodopsin to clone the bovine rhodopsin gene. However, identification of the entire set of photoreceptor-specific genes requires approaches that do not depend on prior knowledge of the components. Suitable methods include subtraction cloning (Swaroop et al. 1991; Rodriguez and Chader 1992) differential hybridization (Bascom et al. 1992; Bernstein et al. 1995), differential display (Liang and Pardee 1992), and representational difference analysis (Lisitsyn et al. 1993; Hubank and Schatz 1994).
In earlier work, Bascom et al. (1992) utilized a differential hybridization screen of retinal cDNA clones to identify genes likely to be important in retinal biology and found ROM1, a photoreceptor membrane protein that localizes to disk rims, and Chx10 (Liu et al. 1994), a retinal homeobox gene. To identify additional retinal-specific genes, we took a similar approach, with a modification aimed at enhancing detection of cone-specific genes, to screen 3717 retinal cDNA clones. Clones that hybridized exclusively or preferentially to a cDNA probe made from the retina of the 13-line ground squirrel [Citellus tridecemlineatus (13GS)], as compared to a probe made from human fibroblast cDNA, were selected for further investigation. By using the 13GS, which has been determined to have a cone-predominant retina (∼95% cones, 5% rods) by morphological, electrophysiological, and molecular criteria (Green and Dowling 1975; West and Dowling 1975; von Schantz et al 1994) we hoped to increase the likelihood of detecting cone-specific genes in the screen.
The 13GS retina probe identifies abundant, evolutionarily conserved sequences whereas the human fibroblast probe identifies sequences derived from ubiquitously expressed genes. Genes that are evolutionarily conserved and expressed at high levels in a tissue-specific fashion are likely to be important for that tissue’s unique identity and function. From the original 3717 clones, we selected 63 for further study based on their preferential hybridization to the 13GS retina probe and ultimately obtained sequence information on 45 of the 63. Nine (20%) of these 45 cDNAs were identified as retinal-specific, validating the utility of this screening method in identifying retinal-specific genes.
RESULTS
Hybridization of 13GS Retina RNA to Cone and Rod cDNAs
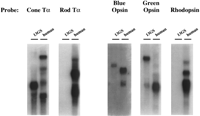
To determine whether utilization of a 13GS retinal cDNA probe for the differential hybridization screen would increase the odds of finding cone-specific genes, we first compared the hybridization of human cDNAs from the cone and rod versions of known cone/rod gene pairs to 13GS retinal RNA. As shown in Figure 1, Northern blots of 13GS and human retinal RNA probed with the human cone transducin α subunit (Tα) and human cone opsins (blue and green) show roughly equal hybridization of these probes to ground squirrel and human retinal RNA. In contrast, human rod Tα and rhodopsin (the rod opsin) hybridize strongly to human retinal RNA and almost undetectably to 13GS retinal RNA. Thus, for these two cone/rod cDNA pairs, 13GS retina RNA hybridizes almost exclusively to the cone version. Similar results were obtained by von Schantz and colleagues (1994). Although we cannot rule out the possibility that the human rod Tα and rhodopsin cDNAs do not hybridize well to 13GS retina RNA because of species variation in the orthologous genes, these results are consistent with the morphologic and electrophysiologic evidence indicating that the photoreceptors of the 13GS retina are mainly cones and suggest that utilizing the ground squirrel as a source of retinal RNA for the differential hybridization screen will increase the likelihood of identifying cone-specific genes.
13GS retina preferentially hybridizes with the cone version of cone/rod cDNA pairs. Northern blots of total RNA (10 μg/lane) from 13GS and human retinas were probed with the cone or rod version of known rod/cone gene pairs. Blots were hybridized with 1 × 106to 2 × 106 cpm/ml of the appropriate probes. Exposure times: Tα cone, 8 days; Tα rod 16 hr; blue opsin, 11 days; green opsin, 3 days; rhodopsin 2.25 hr.
Optimization of Differential Hybridization Screening Conditions
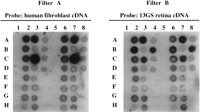
To determine the optimal hybridization and wash conditions for our differential hybridization screen, we utilized a panel of test clones that included some known photoreceptor-specific cDNAs (rhodopsin, recoverin, green opsin) as well as some cDNA clones from genes expressed in the retina as well as elsewhere (ornithine-δ-aminotransferase and others). We maintained a constant hybridization temperature of 42°C and varied the formamide concentration in the hybridization solution from 30% and 50% to give a range of hybridization stringencies. We also maintained the salt concentration in the wash buffer constant at 2× SSC but varied wash temperatures between 42°C and 65°C to provide a range of stringencies for the wash step. Empirically, we chose the conditions (hybridization in 30% formamide, 1 m NaCl, 1% SDS, 10% dextran sulfate at 42°C overnight, washing in 2× SSC, 1% SDS, three times for 20 min at 65°C) under which the known retinal cDNA clones gave the most preferential hybridization to a total cDNA probe prepared from RNA isolated from 13GS retina as compared to a total cDNA probe prepared from RNA isolated from cultured human fibroblasts. Figure 2 shows duplicate lifts of the test panel probed and washed under optimal conditions.
Hybridization of test grids under optimal conditions. Duplicate filters with an array of control phage cDNA clones (see chart
below for clone identity) were probed with human fibroblast cDNA (filter A) or 13GS retina cDNA (filter B) and washed under
the conditions in which the known retinal clones give the best preferential hybridization to the 13GS probe. Recoverin (B4
and B8) exhibits the greatest intensity difference between the two probes. Optimal conditions were determined to be hybridization
in 30% formamide, 1 m NaCl, 1% SDS, and 10% dextran sulfate at 42°C, washing in 2× SSC, 1% SDS, three times for 20 min at 65°C.
 *From E.L. Germaine-Lee, C. Obie, and D. Valle (in prep.).
*From E.L. Germaine-Lee, C. Obie, and D. Valle (in prep.).
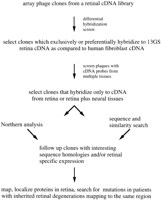
Differential Hybridization Screen of Retinal cDNA Clones
The steps of our screening strategy are outlined in Figure 3. Briefly, duplicate arrays of clones from a human retinal cDNA library were hybridized with a total cDNA probe prepared from 13GS retina RNA and with a total cDNA probe prepared from human fibroblast RNA. Clones that hybridized preferentially to the 13GS retina probe were selected for further study. We subjected the primary screen positives to a secondary screen, which we call multitissue plaque-lift screening. We arrayed the clones and took several lifts from each plate, which we then screened with total cDNA probes from various human tissues (cerebellum, fibroblast, kidney, liver, adrenal gland, and retina). Clones that hybridized to the total cDNA probe from any tissue other than cerebellum and retina were discarded from further investigation. In our experience, multitissue plaque-lift screening is not as sensitive as Northern blot analysis on individual clones, but it provides a quick method for surveying a large number of clones. Finally, we obtained sequence information for clones surviving the secondary screen and used this sequence to search the public databases.
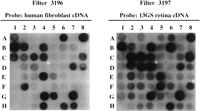
In an initial pilot screen of 945 clones from a retinal cDNA library, we selected 22 clones based on their preferential hybridization (11 strongly positive, 11 positive) to the 13GS retina probe as compared to the human fibroblast probe. Figure 4 shows examples of clones selected for further study. Twenty of these were recoverable from the original phage stocks. In this pilot study, we omitted the multitissue plaque-lift screening step and obtained sequence information from all of the clones (except one, because of an inability to PCR-amplify the insert) to determine the types of clones the screen detected. Sequence data from each clone were used to search the public databases [BLASTN, BLASTX 1.4.9MP (Altschul et al. 1990)]. Among the 19 clones from which sequence was obtained, we identified cDNAs from two photoreceptor-specific genes, interphotoreceptor retinoid binding protein and rod arrestin (two clones), and three that encode neural-specific isoforms of enzymes (two aldolase C, one creatine kinase). Table 1 summarizes the search results for the 19 clones from this pilot screen.
Sample differential hybridization results. Duplicate filters with an array of 63 unknown clones from a human retina cDNA library in λgt10 were probed with human fibroblast cDNA (filter 3196) or 13GS retina cDNA (filter 3197). Clones that hybridized exclusively or preferentially to the 13GS retinal probe were selected for further investigation. Two examples of clones marked for further study, D4 and F8, are indicated by white asterisks.
Summary of Clones Isolated in the Pilot Differential Hybridization Screen Study
Encouraged by the results of our pilot study, we went on to screen an additional 2772 clones. We selected 45 for further study on the basis of their preferential hybridization to the 13GS retina probe as compared to the human fibroblast probe (8 strongly positive, 37 positive). Forty-three were recoverable from the original phage stocks. To minimize the number of nonretinal-specific clones sequenced, we subjected these 43 clones to the secondary multitissue plaque-lift screen. This step eliminated 13. We obtained sequence data on 26 of the remaining 30 clones and used these sequences in searches of the public databases [BLASTN, BLASTX 1.4.9MP (Altschul et al. 1990)]. Table 2summarizes our data on these 26 clones. Six are photoreceptor-specific; eight are neural-specific, four encoding neural-specific isoforms of enzymes, and four encoding proteins involved with synaptic transmission. Additionally, we performed Northern blot analysis on nine clones that detected no significant homologies with sequences in the public databases to identify those of interest because of a retinal-specific expression pattern (data not shown). This screening identified two clones (32 and 39) that were selected for further study because their expression was limited to retina and brain. Both clones detected a transcript of ∼2.3 kb in these tissues, and subsequent sequence analysis has revealed they derive from the same gene.
Summary of Clones Isolated in the Large-Scale Differential Hybridization Screen
The sequence tags of cDNAs identified in this screen and not already in GenBank have been deposited as retinal ESTs and their accession numbers are provided in Tables 1 and 2.
DISCUSSION
Despite recent progress, there are many inherited retinal disorders for which the responsible gene has yet to be identified (Rosenfeld et al. 1994). Among those whose molecular etiology is known, several are caused by defects in retinal-specific genes. For example, six of the eight known retinitis pigmentosa genes encode components of the visual transduction cascade or structural components of photoreceptors (Dryja and Li 1995). Thus, the identification of additional retinal-specific genes will provide strong candidates for the “orphan” inherited retinal diseases, as well as provide reagents for the study of normal retinal biology. In humans, the identification of cone-specific genes is particularly challenging because of the relatively small fraction of cones (∼5% of all photoreceptors) but also particularly rewarding because of the key role of cones in visual function. We devised a differential hybridization screening strategy to identify novel retinal-specific genes with an emphasis on cone-specific genes.
We first assessed the utility of RNA from the cone-rich 13GS retina as a tool for increasing the likelihood of identifying cone-specific cDNAs. We demonstrated that 13GS retinal RNA hybridizes preferentially with cDNAs from the cone Tα subunit and cone opsins as compared to their rod counterparts. This result validates the use of 13GS retinal cDNA to enhance detection of cone-specific cDNAs and provides additional molecular evidence, in support of the morphological, electrophysiological, and existing molecular data (von Schantz et al. 1994) that the 13GS retina is cone-rich. We then modified a technique used in earlier work (Bascom et al. 1992) to perform a differential hybridization screen of 3717 clones from a human retinal cDNA library to identify retinal-specific clones. Our approach involves screening gridded arrays of retinal cDNA clones with a human fibroblast cDNA probe and a 13GS retina cDNA probe. The 13GS retina probe identifies abundant, evolutionarily conserved cDNAs that are expressed in retina, whereas the fibroblast probe identifies cDNAs from genes expressed in multiple tissues. Those clones that hybridized to the 13GS retina probe, but not the human fibroblast probe, are selected for further investigation as candidates from genes likely to be important in photoreceptor health and function.
Although Northern blot analysis showed hybridization of full-length human blue and green opsin cDNAs to 13GS retinal RNA, the human blue and green opsin clones in the test panel do not preferentially hybridize to the 13GS retina probe. This may be attributuable to the relative abundance of green and blue opsin cDNAs in the 13GS retina probe or to weak conservation over the portion of the cDNAs contained in these individual clones. We did not investigate this issue further. However, it demonstrates that the screening method will not necessarily identify every clone of interest.
From the original 3717 cDNA clones screened, we selected 67 (63 recoverable) for further investigation and obtained partial sequence information on 45 of the 63 clones. Nine of these clones (20%) are from known retinal specific genes (Table 3), including two cDNAs for X-arrestin (C-arrestin), the cone arrestin (Murakami et al. 1993; Craft et al. 1994). Additionally, 7/45 (15.6%) clones are from neural-specific enzymes and 4/45 (8.9%) are from genes involved in synaptic transmission (Table 4). Northern blot analysis of the nine clones with no significant matches in the databases has revealed two others of interest because of a retinal- or neural-specific expression pattern. Although we found only two cone-specific clones (both for X-arrestin), the relatively small number of cDNAs screened thus far makes it difficult to assess whether this differential hybridization screening strategy will actually be useful for identifying cone-specific genes. Regardless, these numbers demonstrate that the strategy is successful in identifying cDNAs important in photoreceptor function. Recently developed microarray technology (Schena et al. 1995) will allow upscaling of this method, facilitating rapid detection of differentially expressed retinal transcripts.
Known Photoreceptor-Specific Genes Found in the Differential Hybridization Screen
Some Neural cDNAs Found in the Differential Hybridization Screen
METHODS
Plasmids
Plasmids pB118AS (human rod transducin) and pB117A1 (human cone transducin) were kindly provided by James Hurley (University of Washington, Seattle). Plasmids pSM129 (blue opsin) and hs7 (red opsin) were kindly provided by Jeremy Nathans (The Johns Hopkins University School of Medicine, Baltimore, MD).
Probe Preparation
To make total cDNA probes for the differential hybridization screen and for multitissue plaque-lift screening, total RNA was isolated by guanidinium thiocyanate extraction (cultured human fibroblasts, 13GS retina, human retina, and human cerebellum) (Chirgwin et al. 1979) or purchased from Clontech (human adrenal, kidney, and liver). First-strand cDNA was then reverse transcribed using oligo(dT) as a primer, and the second strand was synthesized and labeled using random hexamer primers with incorporation of [32P] dCTP (modified from Sambrook 1989 Sambrook et al. 1989)]. Probes for the 13GS versus human retina Northern blots were made by excising inserts from the appropriate plasmids or, in the case of rhodopsin, PCR-amplifying the insert from a λgt10 phage clone (kindly provided by Jeremy Nathans) and labeling the inserts according to the method ofFeinberg and Vogelstein (1983).
Differential Hybridization Screen
Duplicate plaque lifts of arrays of well-isolated phage clones from a retinal cDNA library (Bascom et al. 1992) were hybridized (one with human fibroblast probe, one with 13GS retina probe) with 106 cpm/ml of probe in 30% formamide, 1 m NaCl, 1% SDS, and 10% dextran sulfate at 42°C overnight. Probes were preannealed with human placental DNA prior to hybridization, as described (Geraghty et al 1993) to block repetitive elements. Filters were washed in 2× SSC, 1% SDS, three times for 20 min at 65°C.
Multitissue Plaque-Lift Screening
Arrays of phage clones selected from the differential hybridization screen were hybridized with total cDNA probes made from various human tissues (adrenal, cerebellum, fibroblast, kidney, liver, retina). Hybridization and wash conditions were the same as for the differential hybridization screen.
Northern Blot Analysis
13GS retina and human cerebellum, fibroblast, retina, and retinoblastoma cell line (weri-1, weri-27) RNAs were isolated by guanidinium thiocyanate extraction (Chirgwin et al. 1979). Human kidney and liver RNAs were purchased from Clontech. Northern blotting was performed as described (Brody et al. 1992).
Sequencing
Inserts from phage clones were PCR-amplified using phage arm-specific primers (5′- CCACCTTTTGAGCAAGTTCAG-3′, 5′-GAGGTGGCTTATGAGTATTTC-3′). The products were electrophoresed on agarose gels, and the fragments visualized with EtBr and extracted from the gel (Qiagen Gel Extraction Kit, Gene Clean Kit, or NACS columns). The products were then sequenced using a Sequenase (U.S. Biochemical, Inc.) kit following denaturation for 5′ at 95°C. Some inserts were cloned using a TA cloning kit (Invitrogen), and the plasmids were sequenced (U.S. Biochemical, Inc.) with vector primers (Sp6, T7).
Acknowledgments
We thank Sandy Muscelli for preparation of the manuscript. This work was supported in part by the Foundation Fighting Blindness (D.V.), the Milton and Ruth Steinbach Foundation (D.V.), Networks of Centres of Excellence of Canada (R.M.), and the RP Eye Research Foundation of Canada (R.M.). D.S. and S.X. are students in the Predoctoral Training Program in Human Genetics (5T32GM07814). D.V. is an Investigator in the Howard Hughes Medical Institute. R.M. is an International Research Scholar in the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵5 Corresponding author.
-
E-MAIL david.valle{at}qmail.bs.jhu.edu; FAX (410) 9557397.
-
- Received January 2, 1997.
- Accepted February 24, 1997.
- Cold Spring Harbor Laboratory Press