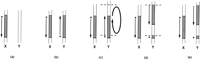Evolutionary Features of the 4-Mb Xq21.3 XY Homology Region Revealed by a Map at 60-kb Resolution
- 1Department of Molecular Microbiology and Center for Genetics in Medicine, Washington University School of Medicine, St. Louis, Missouri 63110; 2Division of Bone and Mineral Diseases, Washington University School of Medicine, The Jewish Hospital of St. Louis, St. Louis, Missouri 63110; 3J.C. Self Research Institute of Human Genetics, Greenwood Genetic Center, Greenwood, South Carolina 29646
Abstract
Forty-three yeast artificial chromosomes (YACs) from the X chromosome have been overlapped across the 4-Mb Xq21.3 region, which is homologous to a segment in Yp11.1. The region is formatted to 60-kb resolution with 57 STSs and is merged at its edges with contigs specific for X. This allows a direct comparison of marker orders and distances on X and Y. In addition to some sequence variation and possible differences in marker order, two larger evolutionary divergencies between the X and Y homologs were revealed: (1) The X homolog is interrupted by a small X-specific region detected by a 3-kb plasmid probe for locus DXS214. An STS was developed from one end of the probe, but the sequence at the other end was highly homologous to an L1 repetitive element. This suggests that the interpolation of the X-specific segment may have involved an L1-mediated event. (2) A 250-kb portion containing DXYS1 is several megabases away from the rest of the homologous DNA on the Y but is contiguous with the remainder of the homologous region on X. Marker orders are consistent with the origin of the Y-specific 250-kb region in a paracentric inversion after the initial transfer of X DNA to the Y chromosome.
[All sequence data for the STSs are in the Genome Database and on the Washington University web site at http://genome.wustl.edu/cgm/cgm.html.]
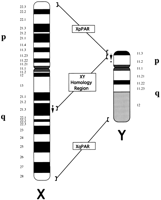
In addition to several smaller regions (Affara and Ferguson-Smith 1994), three major regions of homology are shared by the human X and Y chromosomes (Fig.1). Two of them comprise the pseudoautosomal regions at the tips of the p and q arms, respectively 2.5 and 0.35 Mb in length. The other homologous region, in Xq21.3 and Yp11.1 (Foote et al. 1992; Vollrath et al. 1992; Affara and Ferguson-Smith 1994), differs from the pseudoautosomal segments in several respects. First, it is more extensive, on the order of 4 Mb. Second, unlike the pseudoautosomal portions, the homologous X and Y segments are not thought to pair during meiosis. In the chimpanzee and lower vertebrates, this region only exists on the X chromosome (Page et al. 1984; Geldwerth et al. 1985; Koenig et al. 1985; Affara and Ferguson-Smith 1994). Hence, the XY homology region is unique to humans and presumably arose by translocation of the region from the X to the Y chromosome relatively recently on the evolutionary time-scale.
A clone map of the XY homology region on both X and Y can help to determine features of the evolution of the sex chromosomes and to initiate a comparison of their genetic content. Most of the euchromatic region of the human Y chromosome has been cloned in yeast artificial chromosomes (YACs) and formatted with sequence-tagged sites (STSs) (Foote et al. 1992; Vollrath et al. 1992). A more recent map of the Y chromosome is provided by Jones et al. (1994), although marker order within the XY homology region is essentially the same as in Foote et al. (1992). On the Y chromosome, the corresponding DNA includes STSs sY20, sY21...sY52, in numerical order across a 4-Mb region, and a relatively small segment containing DXYS1, located ∼2 Mb more proximal.
The homologous region on the X chromosome has been less well characterized. Starting with the sY STSs from the Y chromosome map and supplementing them with 32 additional STSs developed from YAC insert ends, we have developed a YAC/STS contig that encompasses the entire homology region of >4 Mb, reaching X-specific DNA at both ends. Direct comparison of marker order and spacing suggests some features of the evolution of this uniquely human XY homology region.
RESULTS
Construction and Internal Consistency of the Contig
Some information about the placement of a group of XY homologous markers in Xq21 had been derived from fluorescent in situ hybridization studies, and other ordering information had been inferred using a panel of deletion breakpoint hybrids, derived primarily from DNA of choroideremia patients (Page et al. 1984; Geldwerth et al. 1985; Koenig et al. 1985; Philippe et al. 1993). From such data, successive bins were found to include sY73 (DXYS1) and sY20 (DXYS42); sY24 (DXYS5); DXS214; and sY48 (DXYS12). It was thus clear that sY73 had a different relative location on X compared to Y (where it was on the other side of sY48). We therefore determined to make an X-specific map without regard to the published order on the Y and then to compare the resulting maps.
Contig construction was initiated by screening the series of sY STSs against several YAC libraries to make small contigs. New STSs were then developed from YAC insert ends for recursive screening to achieve long-range contiguity across the entire region (Kere et al. 1992), with three- to eightfold clone coverage. Four additional STSs were placed in the contig, including DXS214 (discussed below); DXS1217 (AFM288ye9), a polymorphic marker in the X-specific region at the centromeric end of the contig (Fig. 2); XSTS0460, an X chromosome STS derived from an X DNA clone; and DXS3 (Stanier et al. 1991).
Major regions of homology between the human X and Y chromosomes. For the XY homology region, the asterisk (*) and the arrow represent marker order differences between the X and Y by comparison of the physical map of the Y chromosome (see text) to the map of the X chromosome presented here. The asterisk represents the region surrounding DXYS1, and the arrow represents the general order of 33 markers (sY20–sY52). (XpPAR) Xp pseudoautosomal region; (XqPAR) Xq pseudoautosomal region.
YAC/STS map of the Xq21 XY homology region. (a) Markers, including X-specific markers (DXS1217, DXS214, and DXS3) and XY homologous markers corresponding to sY markers shown below. (b) Megabase scale. (c) Brackets show corresponding regions of homology on the Y chromosome, including those that are Yp proximal (represented by asterisks in Figs. 1 and 3) and Yp distal (represented by arrows in Figs. 1 and 3). (d) Differentially shaded bar represents X-specific regions (light shading) and XY homologous regions (dark shading). (e) Order of markers, identified by common names, from centromeric to telomeric. Markers shown as a number followed by an L or R identify YAC-end STSs; the number is the yWXD Washington University database accession number of the YAC; L or R indicates the left or right end of the YAC. XSTS0460 is a random X-specific marker. (f) YACs and STS content. YACs are drawn to scale and are identified by a yWXD number followed by either a letter identifying the library of origin (E or F libraries) or the complete library location [(I) ICI library; (M) CEPH mega-YAC library]. (○) The origin of the YAC-end STSs; (•) STS content in the YACs. Several STSs were spaced too closely to be discriminated from neighboring STSs and have not been illustrated on the map. These include sY30, sY31, sY33, sY34, sY36, and sY37, which have been placed in general numeric order in relation to the other sY markers. The marker sY25 could not be placed accurately on the map because of weak signals from X-specific templates but is placed tentatively between sY24 and sY26 (see text). The size of the X-specific region at DXS214 is unknown and is not necessarily drawn to scale.
In the completed contig (Fig. 2), the STS content of all the YACs is shown, and all YACs are drawn to scale. This provides estimates of the size of the complete region and of spacing between markers. The XY homology region spans 4 Mb of the 4.5 Mb in Figure 2. At the centromeric end, a 200-kb X-specific region is shown that is merged with a published 3-Mb contig spanning the candidate region for cleft lip and palate disease (CPX) (Forbes et al. 1996). At the telomeric end of the contig, the 150-kb X-specific region shown is merged with overlapping YACs that span the remainder of Xq21.3 and Xq22 (Vetrie et al. 1994; A.K. Srivastava, M. Shomaker, C. Jermak, S. Mumm, and R. Nagaraja, in prep.). Hence, the entire XY homology region is clearly covered by this contig.
In general, only a few YACs recovered from this region were chimeric and were dropped from the map of Figure 2. Consistent with this index of quality, end clones from even the largest YACs mapped to the expected locations, and the STS content in the YACs was also totally consistent, with no indications of internal deletions. Thus, in Figure2, the lengths of the 43 YACs and the inter-STS distance of 60 kb are relatively reliable.
Features of the X Chromosome That Vary from the Y Homolog
All of the STSs derived from X-specific YAC ends that are contained within the boundaries of the XY homology region were XY homologous. This inference is based on the comparable amplification of the STSs from human/hamster hybrid cell lines containing either the human X or Y chromosome. During the mapping, however, it became increasingly clear that not all XY homologous STSs amplify equally well from X- and Y-specific templates. Among the sY primers that showed consistent differential amplification, the most striking example was sY25, which gives robust signals from Y DNA but produces only weak signals from a variety of X-specific DNA preparations. Consequently, this STS could not be used to screen libraries for X YACs and was placed on the map by testing clones and reamplifying signals. Differential amplification is consistent with previous Southern analyses, suggesting some sequence differences between the X and Y homologs (see Discussion).
One X-specific marker, DXS214, was shown previously to lie within the boundaries of the XY homology region, between the markers sY24 and sY48 (Philippe et al. 1993). To localize the marker on the map, an STS was developed. Both ends of the insert for the probe pPA20 (DXS214) were sequenced. CENSOR (Jurka et al. 1996) analysis showed that one end has high homology to L1-type repetitive elements. Sequence from the other end showed no matches to repetitive elements using CENSOR with default parameters, and an STS was developed. The STS showed high background at lower annealing temperatures but was specific enough to screen for cognate YACs when tested at an annealing temperature of 60°C. DXS214 was thereby placed between sY42 and sY43.
DISCUSSION
Chromosomal Rearrangements Between X and Y
When marker order and distances are compared, divergence is revealed at several levels of organization. The largest-scale difference between the X and Y homologs is the location of DNA in and around the DXYS1 (sY73) locus. From the physical maps of the Y chromosome (Foote et al. 1992; Jones et al. 1994), most of the homologous DNA is physically contiguous, containing STS markers ordered by number from sY20 to sY52; but a smaller fragment containing DXYS1 is several megabases more proximal. In contrast, for the X chromosome (Fig. 2), DXYS1 and sY20 are closer together. Both are included in three YACs from different libraries. The distance between DXYS1 and sY20 must be <300 kb (the size of yWXD5264) and is more likely on the order of 100–200 kb. This is consistent with contiguity for the DNA containing DXYS1 and the rest of the XY homology region at the sY20 end.
The DXYS1-containing region on the Y chromosome had a previously estimated minimal size of 36 kb; no maximum size range was determined (Page et al. 1984). More recently, the maximum size was established at 280 kb (Sargent et al. 1996). From our map, the likely size of this region is on the order of 250 kb. An additional four STSs proximal to sY73 on the X chromosome would presumably move with DXYS1 to the displaced segment on the Y, though this remains to be confirmed.
The other most obvious difference between the X and Y homologs is the presence of additional DNA in the middle of the X region, containing the X-specific probe DXS214. The STS developed for this marker places it between sY42 and sY43. One end of the probe for DXS214 has high sequence homology to L1 repetitive elements; the other end appears to have unique sequence. The entire probe pA20 is ∼3 kb, but the X-specific region surrounding DXS214 could be much larger.
Concerning the origin of the X-specific DXS214 region, it might have existed on the X chromosome when the initial transfer was made to Y and was then deleted from the Y chromosome. Alternatively, the region might have been inserted into the X chromosome after the initial transfer from X to Y. In either case, the presence of the L1 element in DXS214 suggests that such an element may have been involved. Sequencing of the region and identification of the X-specific/XY homologous boundaries may provide further indications of a possible underlying mechanism.
Apart from major rearrangements, the order of markers on the X chromosome is in agreement with the binning of five markers in hybrids containing DNA from patients with X-to-autosome translocations (Philippe et al. 1993) and generally follows the order sY20–sY52 reported for the Y chromosome. There are differences, however, one significant one and two minor ones. sY50 is placed between sY43 and sY44 on the X chromosome, at a considerable distance from its position between sY49 and sY51 on the Y chromosome maps. Because the locations are supported by several independent YACs, they suggest another potential rearrangement that has arisen since the transfer of the homologous region from X to Y. In addition, the order of two pairs of markers (sY46 and sY47, sY48 and sY49) are apparently reversed on X compared to Y. The resolution and clone coverage of the two maps is somewhat different, however, so that further work will be required to determine whether these apparent differences are intrinsic to uncloned DNA of the X and Y chromosomes.
During the preparation of this manuscript, a set of four contigs covering portions of the XY homology region was published (Sargent et al. 1996). The separate contigs (0.2, 0.5, 1.5, and 1.2 Mb) total 3.4 Mb of the XY homology region. The orientation of these contigs and the order of markers were based on the X chromosome deletion panel described by Phillipe et al. (1993). The map presented here extends the analysis by providing uninterrupted clone coverage of the entire region. Thus, gaps are filled, totaling >0.6 Mb, and both ends of the XY homology region are merged with neighboring chromosomal DNA. Furthermore, these results are based solely on STS content of YACs, so that the contig alignment determined by deletion panels is an independent verification rather than the sole source of order. The provision of complete DNA coverage, in turn, allows for an accurate estimate of the 4-Mb extent of the region and for comparison of marker order on the X and Y chromosomes. Consequently, it is clear that the marker sY73 (DXYS1) is contiguous with the remainder of the XY homology region on the X chromosome, with no other intervening X-specific material; and by comparison to the maps of the Y chromosome, that sY73 (DXYS1) is separate from the remainder of the XY homology region (Foote et al. 1992; Vollrath et al. 1992; Jones et al. 1994).
At a finer sequence level, comparisons between the X and Y chromosomes in the region have revealed 98% identity for the probe St25 (Koenig et al. 1985) and >99% for DXYS1 (Page et al. 1984). The ability of the STSs across the region to amplify from X or Y is consistent with high overall homology (98%–99%) across the region. From our results, however, the difference in efficiency of amplification from X and Y templates with some STSs is an indication of possibly greater local sequence variation. Preliminary direct sequence analysis for STSs in this region shows levels of identity for X and Y ranging from 96% (for sY25) to 100% (for sY36, sY51) (S. Mumm and D. Schlessinger, in prep.). It will be interesting to determine whether the most highly conserved regions are constrained by coding or other functions.
Evolution and Possible Gene Content of the XY Homology Region
High sequence homology is consistent with the notion of an evolutionarily recent transposition of this region from the X to the Y chromosome. The sex chromosomes would then have diverged somewhat, with the divergence maintained by the absence of chiasmata in this region during male meiosis. Based on the initial common probe content (Page et al. 1984; Geldwerth et al. 1985; Koenig et al. 1985), the physical map of the Y (Foote et al. 1992; Vollrath et al. 1992; Jones et al. 1994), and now the physical map of the X, Figure 3 shows a schematic of the possible course of a potential mechanism for the divergence of X and Y in the region. The presence of the XY homology region solely on the X chromosome of apes indicates its origin as X-specific on the homonid ancestor of apes and humans (Fig. 3a). At some point after divergence of apes and humans, the region was duplicated and transposed from Xq21.3 to Yp11.1 in the human lineage (Fig. 3b). A paracentric inversion on Y would have separated DXYS1 from the rest of the region, and changes in either X or Y could have led to the discordant positions of sY50, and so forth (Fig. 3c,d). The X-specific region at DXS214 could then be accounted for by an insertion into the X chromosome or a deletion from the Y, after the original transposition of the region from the X to the Y chromosome (Fig. 3e). The presence of DXS214 and the displacement of DXYS1 on Y chromosomes in all human populations suggest that those changes likely arose before a bottleneck in human evolution fixed them in the genome.
Possible events in the evolution of the XY homology region. (a) Structure of the X and Y chromosomes of the human precursor, where the region is unique to the X chromosome at Xq21. As in Figs. 1 and 2, the asterisk (*) represents the region surrounding DXYS1 and the arrow represents the general order of markers sY20–sY52. (b) This region was then transposed to the Y chromosome at Yp11. (c) A paracentric inversion occurred on the Y chromosome to generate the structure in d, where the DXYS1 region was separated from the remainder of the XY homology region. (e) The X-specific region at DXS214 originated as an insertion into X or a deletion from Y, potentially via an L1-mediated mechanism (see text).
The availability of the complete physical maps of both X and Y should facilitate the systematic analysis of possible gene content and comparative structure. Although no genes from the XY homology region have been reported yet, we believe the area to be transcriptionally active. Several of the YAC-end STSs generate signals by RT–PCR and are positive on top-level pools from cDNA libraries. In particular, sWXD1358 is positive by RT–PCR and from pools for a teratocarcinoma cDNA library (S. Mumm and M. D’Esposito, unpubl.). Because human males, compared to other primate males, have this region on both X and Y, they could have twice the dosage of any constituent genes. If dosage compensation were important for such genes, they would become possible candidates for involvement in Turner syndrome-like pathology (Zinn et al. 1993). Genes in the XY homology region may also be candidates for premature ovarian failure with breakpoints in Xq21 (Forabosco et al. 1979).
METHODS
STSs
The set of STSs developed from the Y chromosome were obtained from David Page (Vollrath et al. 1992). STSs from YAC ends were made as described in Kere et al. (1992) and use either TNK50 or TNK100 buffer as stated in Table 1 (Blanchard et al. 1993). The STS for DXS3 was described in Stanier et al. (1991). Marker DXS214 (pPA20) was kindly provided by Christophe Philippe and Frans Cremers (University Hospital Nijmegen, The Netherlands). An STS for DXS214 was developed by partially sequencing pPA20 using cycle sequencing (Srivastava et al. 1992). Both ends of the insert were sequenced using pBR322 HindIII forward and reverse primers. STS primers for DXS214 were designed using the OSP program (Hillier and Green 1991). All relevant STS information is available in the Genome Data Base and via the Genome Center World Wide Web page (http://genome.wustl.edu/cgm/cgm.html).
STSs Developed in This Study
YACs and Screening
YACs were obtained from a variety of libraries, including the E library (Nagaraja et al. 1994), the F library (Lee et al. 1992), the Zeneca, Inc. library (Riley et al. 1990), and the Centre d’Etude du Polymorphisme Humain (CEPH) library (Albertsen et al. 1990). All libraries were screened with STSs, and the cognate YACs recovered were sized by pulsed-field gel electrophoresis. YACs are described with additional information in the Genome Data Base as well as the Genome Center web page; all are available from the American Type Culture Collection (ATCC).
To ensure that the clones in the contig were from the X chromosome, the libraries initially screened were all derived from female DNA. Some screenings were also done with one collection from a male source, the CEPH mega-YAC collection; but care was taken to recover only X chromosome-specific YACs (e.g., by screening with the X-specific probe DXS214).
Mapping started by using the sY primers to isolate a contingent of clones and then determining the content of neighboring STSs in colony-purified YACs. Additional STSs from YAC end inserts were then used in further screenings to “walk” to overlapping clones until subcontigs merged across the entire region. Markers DXS1217, XSTS0460, and DXS3 were screened as STSs in the Genome Center and were incorporated into the XY homology region when they identfied cognate YACs already in the contig.
Acknowledgments
We thank Sandra MacMillan for handling and storing clones and STS reagents, and for sizing YACs; and Frank Burough for CENSOR runs. S.M. was supported by National Institutes of Health (NIH) Training grant 5T32 AR07033-21 and a research fellowship from the Shriners Hospitals for Crippled Children. Further support was provided by GESTEC grant HG00201 (NIH).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL davids{at}sequencer.wustl.edu; FAX (314) 362-3203.
-
- Received September 16, 1996.
- Accepted February 10, 1997.
- Cold Spring Harbor Laboratory Press