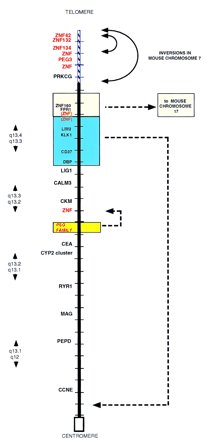
Positions and borders of the major evolutionary rearrangements between human 19q and mouse chromosome 7. Comparative map of human chromosome 19q and related regions in the mouse. The physical map of human 19q is represented, showing the positions of segments that are rearranged in order in mouse. Only a few anchor markers are presented for simplicity. Each crosshatch bar in the map represents a distance of 1 Mb in the human physical map. Colored boxes surround regions that are transposed in order in mouse relative to human; broken arrows indicate the positions to which they have been moved in the mouse genomes. Blue hatching on the backbone of the map indicates the extent of a large inversion in Mmu7 relative to H19q13.4; a smaller inversion, involving genes related to ZNF132, ZNF134, and related clustered zinc finger gene family members, comprise a second, nested inversion within this larger region. Genes located at the borders of each rearranged segment are indicated in red. A number of zinc fingers containing genes have been identified on chromosome 19q and in related regions of Mmu7; some have been characterized and are indicated by specific locus names (e.g., ZNF134); clusters of other genes that have not been completely sequenced or assigned locus names are indicated simply as ZNF on this map. Data are compiled from published studies (Ashworth et al. 1995;Stubbs et al. 1996; Kim et al. 1997; Shannon et al. 1997); additional information can be obtained from the original references.











