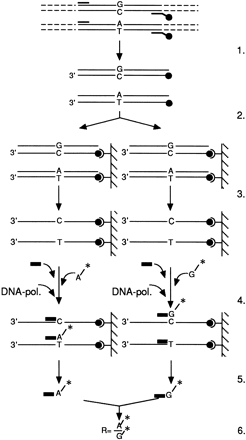
Schematic presentation of the steps in the SPMS method (modified fromSyvänen et al. 1993), mutation 673C → T as an example. (1) PCR with one biotinylated (•) and one unbiotinylated primer. (2) Affinity capture of the biotinylated PCR product on streptavidin-coated microtiter wells. (3) Washing and denaturation. (4) Primer extension reaction. All PCR reactions were aliquoted in eight separate wells. The tritium-labeled nucleotide representing the normal allele (G*) was added in four wells, and four wells contained the tritium-labeled nucleotide representing the mutant allele (A*). Depending on the sequences present, the labeled nucleotide will hybridize next to the minisequencing primer. The nucleotides not incorporated will be washed out. (▪) Detection primer; (A*/G*) [3H]dATP/[3H]dGTP; (DNA-pol.) DNA polymerase. (5) Measurement of the incorporated label. (6) Calculation of the result.











