Interpreting a Sequenced Genome: Toward a Cosmid Transgenic Library ofCaenorhabditis elegans
Abstract
We have generated a library of transgenic Caenorhabditis elegans strains that carry sequenced cosmids from the genome of the nematode. Each strain carries an extrachromosomal array containing a single cosmid, sequenced by the C. elegans Genome Sequencing Consortium, and a dominate Rol-6 marker. More than 500 transgenic strains representing 250 cosmids have been constructed. Collectively, these strains contain approximately 8 Mb of sequence data, or ∼8% of the C. elegans genome. The transgenic strains are being used to rescue mutant phenotypes, resulting in a high-resolution map alignment of the genetic, physical, and DNA sequence maps of the nematode. We have chosen the region of chromosome III deleted bysDf127 and not covered by the duplicationsDp8(III;I) as a starting point for a systematic correlation of mutant phenotypes with nucleotide sequence. In this defined region, we have identified 10 new essential genes whose mutant phenotypes range from developmental arrest at early larva, to maternal effect lethal. To date, 8 of these 10 essential genes have been rescued. In this region, these rescues represent ∼10% of the genes predicted by GENEFINDER and considerably enhance the map alignment. Furthermore, this alignment facilitates future efforts to physically position and clone other genes in the region.
[Updated information about the Transgenic Library is available via the Internet at http://darwin.mbb.sfu.ca/imbb/dbaillie/cosmid.html.]
How is the information in a eukaryotic genome processed to create a living organism with its repertoire of developmental patterns and behaviors? To gain this knowledge first we must be able to recognize a particular sequence of nucleotides as a functional unit, then be able to follow the physical and biochemical workings of this unit, and its products, to their resulting effect on the organism. Toward this goal, the genomes of a number of genetically amenable organisms are being sequenced in their entirety. Of the metazoan genomes presently being sequenced, the complete genomic sequence ofCaenorhabditis elegans is expected to become available to the scientific community first, with an estimated completion date in 1998 (Wilson et al. 1994; Waterston and Sulston 1995). Already, a majority of the gene-rich sequence data have been generated, and from these data an excess of 12,000 protein-encoding genes are predicted (Waterston et al. 1997; S. Jones, pers. comm.). Approximately 50% of these predicted genes share significant sequence similarities with genes of known function. However, such comparative sequence analysis will not provide a complete understanding of how the information encoded in sequence data correlates with biological function (Miklos and Rubin 1996). Other methods, such as the application of transgenesis, will be necessary to elucidate such information. In the case of open reading frames that do not show significant similarity to sequences presently in the databases, mutant analysis is a powerful tool for investigating the biological function of these unique genes.
A large collection of lethal mutants in essential genes exists for many regions of the C. elegans genome: the unc-22 region of chromosome IV (Rogalski et al. 1982; Rogalski and Baillie 1985;Clark et al. 1988); the left portion of chromosome V balanced by the translocation eT1 (Rosenbluth et al. 1988; Johnsen and Baillie 1991); the left portion of chromosome I balanced by sDp2 (Rose and Baillie 1980; Howell et al. 1987; Howell and Rose 1990; McKim et al. 1992; McDowall and Rose 1997a); and chromosome III (H.I. Stewart and D.L. Baillie, unpubl.; M. Edgley, J. Gawne, and A.M. Rose, unpubl.). In some regions, these essential genes have been aligned with the physical map using transgenic rescue.
C. elegans is particularly amenable to transgenesis, which provides a relatively straightforward method of correlating a particular mutant phenotype with a region of nucleotide sequence. By means of cosmid/plasmid transformation rescue experiments (Fire 1986), the phenotype caused by a genetic mutation may be complemented, or “rescued,” by the addition of an extrachromosomal copy of the wild-type gene. Initially, this was done by direct injection into hermaphrodites heterozygous for the allele to be tested. In a region of ∼200 kb immediately to the left of unc-22(IV), the direct injection of six cosmids was used to rescue six genes: let-56and let-653 (Clark and Baillie 1992), let-92 (S.J. Jones and D.L. Baillie, unpubl.); par-5 (D. Shakes, pers. comm.); dpy-20 (Clark and Baillie 1992), and let-60(Han and Sternberg 1990). Once injected into the worm, it is thought that DNA molecules undergo repeated homologous recombination, forming large extrachromosomal arrays containing multiple copies of the injected molecules (Mello et al. 1991). These arrays are thought to be molecularly stable once formed, and a percentage become heritable and are transmitted from generation to generation in a non-Mendelian fashion (Mello et al. 1991). Heritable extrachromosomal arrays can be considered precisely characterized free duplications, and can be moved between strains in the same manner. Hence, a more flexible approach of introducing extrachromosomal DNA into C. elegans entails crossing a genetically marked array, previously constructed in another strain, into a mutant strain of interest (McDowall and Rose 1997b). Although both procedures allow regions of DNA sequence to be correlated with mutant phenotypes, the latter method can be used to test the cloned DNA contained in an array against mutations in different strains, without having to repeat the injection procedure. Previous systematic studies in our laboratory and in others have indicated that an average of one essential gene per cosmid clone can be rescued. This rate of rescue does not necessarily reflect our ability to rescue essential genes, but rather reflects our current ability to saturate a particular region of sequence for essential gene mutations. This approach has the potential to produce high-resolution map alignment, and results in mutant phenotypes being identified for large numbers of predicted proteins.
More than 1500 genetic loci have been identified in C. eleganssince the scientific community first began studying the nematode. Although the consortium has now sequenced the majority of these genes, the actual physical position for most of these genes has yet to be identified. Whereas the relative order of genes along a chromosome is expected to be the same on both the physical and genetic maps, the ratio of physical distance to meiotic recombination distance varies depending on the position of the gene along a chromosome (Barnes et al. 1995). For example, the central gene cluster regions of autosomal chromosomes are reduced recombinationally relative to regions in the chromosome arms. Thus, it is often difficult to predict with accuracy the physical position of a gene, solely on the basis of its genetic map position. As the number of genes with both genetic map positions and positive physical clones increases, the resolution of the map alignment is improved. In this paper we describe a large-scale construction of transgenic strains and their usefulness for comprehensive rescue of mutant phenotypes.
RESULTS
Marking the Transgenic Strains
All transgenic strains created by this project were constructed using wild-type worms (N2 Bristol strain). To facilitate the recognition of transgenic animals in a wild-type genetic background, we used the semidominant mutation of the rol-6 collagen generol-6(su1006) (Kramer et al. 1990) contained in the plasmid pRF4 (Mello et al. 1991). Animals that carry extrachromosomal copies of this mutant rol-6 allele exhibit a helically twisted cuticle and “right-roller” motion, which is easily seen.
As shown by Mello et al. (1991), large extrachromosomal arrays can be formed in vivo when different DNA molecules containing regions of homology are coinjected into the syncytial gonad of an adult C. elegans hermaphrodite. Essentially all C. elegans cosmids used by the Sequencing Consortium are cloned into one of two vector systems, pJB8 or Lorist. Although pRF4 and pJB8 vectors have adequate homology to facilitate the formation of heterogeneous extrachromosomal arrays, minimal homology exists between the pRF4 and Lorist vectors. To increase homology between these two vectors, a kanamycin resistance gene was added to pRF4 to create pCes1943 (S.J.M. Jones and W.B. Barbazuk, pers. comm.). This plasmid was coinjected with all cosmids.
Transgenic Strains
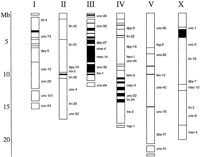
To date, we have constructed transgenic nematode strains for >250 C. elegans cosmids. These cosmids account for approximately 8 Mb of essentially nonoverlapping sequence, including >50% of chromosome III and at least 10% of chromosome IV (see Fig.1). A complete list of cosmids for which transgenic strains are available can be obtained via the internet at the World Wide Web address: http://darwin.mbb.sfu.ca/imbb/dbaillie/cosmid.html. Of the 250 cosmid transgenics reported here, 208 were constructed with purified cosmid DNA obtained directly from the C. elegansGenome Sequencing Consortium. This DNA is an aliquot of that used to generate the sequence data, and hence we have greatly reduced the potential for variation between the published sequences and those of the extrachromosomal arrays generated in our laboratory. For cosmids that the sequencing laboratories were unable to provide DNA, bacterial clones were received from Alan Coulson [Medical Research Council (MRC), Hinxton, UK], and DNA was prepared in our laboratory (see Methods). For these cosmids, the DNA used to create the transgenic strains may differ slightly from that reported by the Consortium, because of clonal variation. As such, cosmid DNA prepared from bacterial clones was used to create transgenic strains only in those regions where purified DNA was not available from the Consortium.
A physical map of the C. elegans genome indicating regions for which we have constructed cosmid transgenic strains. Each of the chromosomes is represented by an open, rectangular box. Approximate physical positions of some genes are indicated by gene name to the right of the chromosome. Filled regions and black lines along the chromosomes represent areas for which transgenic strains are available. Physical markers are placed as per ACeDB [Richard Durbin and Jean Thierry Mieg (1991–) A C. elegans Database. Documentation, code, and data are available from anonymous FTP servers atlirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk, and ncbi.nlm.nih.gov].
For the majority of transgenics constructed in this study, no obviously aberrant phenotypes were noticed. Most strains that carry cosmid arrays segregate only the anticipated Rol-6 and wild-type progeny. This result is not unexpected, as cosmids generally contain the endogenous promoters of most genes present on the cosmid. However, it is possible that the high copy number of genes contained in extrachromosomal arrays could result in the ectopic, overexpression or temporal misexpression of some genes. This aberrant expression was noted by Han and Sternberg (1990) when the cosmid containing the let-60(ras) gene caused a multivulva phenotype when expressed on an extrachromosomal array. In our case, we have observed a small number of cosmid transgenics that appear to have phenotypes not normally associated with transgenesis. Perhaps the most notable of these dominant phenotypes is exhibited by worms that have been transformed with either cosmid C05D2 or F56D2. Normally, spontaneous C. elegans males arise from the progeny of self-fertilizing hermaphrodites at a rate of 0.2% when grown at 20°C (Rose and Baillie 1979). However, transgenic worms constructed from cosmids C05D2 or F56D2 produce males at a rate significantly higher than that expected for wild-type worms (data not shown). As C05D2 and F56D2 overlap by ∼25 kb, it is probable that there is a site common to these two cosmids that results in the overproduction of males when carried in an extrachromosomal array. Currently, we are subcloning these cosmids to determine the nucleotide sequence responsible for this unusual phenotype.
In addition, of the 250 cosmids for which we have created transgenic strains, we have observed six cosmids that do not readily form heritable extrachromosomal arrays under our standard injection conditions. This phenomenon was also noted by A. Fire (1986) who suggested that particular extrachromosomal arrays may cause lethality in worms that carry them. He indicated that this lethality may be attributable to a high dose intolerance of a particular gene product, which may result when an array incorporates more than a few copies of a “poisonous” gene. Although we have successfully constructed heritably transformed lines containing cosmid DNA for all 250 cosmids so far attempted, we found that for six cosmids it was necessary to reduce the relative amount of cosmid DNA used in the injection procedure. It has been suggested by Mello et al. (1991) that changes in the relative DNA concentrations used in the injection mix may alter the ratio of cosmid and plasmid DNA within extrachromosomal arrays. This observation implies that the difficulty we experience in constructing transgenic lines of certain cosmids may be indicative of a concentration/expression threshold limit for those cosmids. Table1 lists the six cosmids for which we were unable to recover transgenic lines at our standard concentration of 80 ng/μl of pCes1943 and 20 ng/μl of cosmid. Shown for each are the lower concentration of cosmid that was used to obtain transgenic lines.
Cosmids for Which Transgenic Strains Could Not Be Constructed Using Standard Injection Conditions
Although it is tempting to predict that specific genes may be responsible for preventing the formation of stable extrachromosomal arrays that contain a high copy number of these “problem” cosmids, it should be noted that this hypothesis is untested. It is possible that the cosmid concentrations used for microinjection were significantly higher or lower than our estimates (see Methods). However, if real, these cosmid-specific phenotypes may provide us with direct biological information that would be difficult to obtain using a purely classic genetic approach.
As reported by Stinchcomb et al. (1985) and Mello et al. (1991), multiple transformed lines obtained from a single injected animal contain extrachromosomal arrays that may differ from each other with respect to heritability, structure, and copy number of cosmid and plasmid DNA. For this reason, we maintain up to three transgenic lines (referred to as segregants) for each cosmid whenever possible.
Confirmation of the Presence of Cosmid DNA in Transgenic Strains
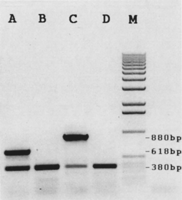
Because cosmid DNA is injected into worms that are genetically wild type, there is no direct phenotypic confirmation of the presence of cosmid DNA, as is often the case when a rescuing cosmid is injected directly into a mutant animal. Therefore, it is possible that some of the extrachromosomal arrays that we recover consist solely of pCes1943 DNA. Hence, it is necessary to confirm the presence of cosmid DNA within extrachromosomal arrays. To do this we used PCR and one of two primer pairs specific for either a portion of the Lorist cosmid vector (McKay 1993) or a portion of the pJB8 cosmid vector. We have confirmed that these primers do not amplify a product from worms transformed by pCes1943 alone (Fig. 2). By performing PCR on DNA derived from all transgenic strains, we are able to confirm the presence of cosmid DNA within extrachromosomal arrays before offering the transgenic strains to the community. From a random sample of 310 transgenic strains that exhibited Rol-6 transmission beyond the F2 generation, 86 did not contain cosmid vector DNA (data not shown), reaffirming the need for PCR testing. As reported by Mello et al. (1991), extrachromosomal arrays are stable once formed. Our results support this observation, as PCR testing confirmed the presence of cosmid vector even after transgenic worms have been allowed to propagate for many generations (data not shown).
A typical agarose gel showing the results of PCR testing of transgenic strains. The 380-bp band in all experimental lanes is the product of the control primers R5 and R11, which were included in all reactions. (Lane A) Results of performing PCR on DNA obtained from an N2 worm that has been cotransformed with a pJB8 vector cosmid and pCes1943. Primers used were DC1 and DC2. (Lane B) Results obtained from an N2 worm that has been transformed with pCes1943 only. Primers used were DC1 and DC2. (Lane C) Results obtained from an N2 worm that has been cotransformed with a Lorist vector cosmid and pCes1943. Primers used are Krp17 and Krp18. (Lane D) Results obtained from an N2 worm that had been transformed with pCes1943 only. Primers used are Krp17 and Krp18. (Lane M) DNA size markers (Promega 1 kb ladder).
To further confirm the integrity and general usefulness of our extrachromosomal arrays, we used the transgenic strains generated by this project to rescue visible mutations in a number of genes, previously placed on the physical map by other researchers. The mutation dpy-17(e164) was shown by A. Smardon to be rescued by the cosmid F54D8. We used our transgenic strain of the cosmid F54D8 to duplicate this result, proving that the extrachromosomal array contained at least the minimum sequence needed to rescue thedpy-17 gene. This same duplication of results was shown forunc-32(e189) (previously positioned to cosmid ZK637 by J. Sulston, Sanger Centre, Cambridge, UK), and sma-3(e491)(previously positioned to R13F6 by C. Savage, Waksman Institute, Piscataway, NJ). All of these crosses were performed using transgenic strains that had been maintained on plates for a year or more. These results support Mello’s theory that extrachromosomal arrays undergo little, if any, internal rearrangements after their initial formation (Mello et al. 1991).
Cosmid Rescue Experiments
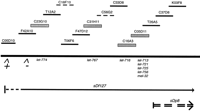
We have used the transgenic strains generated by this project to begin a systematic correlation of the genetic and physical maps in the gene cluster region of the left arm of chromosome III. Although subcloning experiments are often required to locate precisely the sequence responsible for rescuing a particular mutant phenotype, candidate sequences can be quickly narrowed down with cosmid rescue data, using simple genetic techniques and the transgenic strains we have constructed. As a starting point, we chose to concentrate our rescue efforts on genes that could be localized to a reasonably small section of the physical map. The physical position of 22 essential genes, generated in the Baillie laboratory and shown to be deleted by the deficiency sDf127, was determined by localizing the physical end points of the deficiency. Using deficiency PCR, the left and right end points of sDf127 were mapped to the cosmids C23G10 and T20H4, respectively (see Methods). Also using PCR, we determined that the left end point of sDp8 is between cosmids M01G4 and C05D11 (N.J. O’Neil, in prep.). Ten of the 22 essential genes deleted by sDf127 were not complemented bysDp8, placing those genes between cosmids C23G10 and M01G4. Beginning with the transgenic of cosmid C23G10 and working right along the physical map shown in Figure 3, we systematically tested available transgenic strains against mutant strains for each of the 10 essential genes localized to this region. In doing so, we were able to correlate the mutant phenotypes of eight essential genes to particular cosmids, as summarized in Table 2. All of these rescues were full rescues; therefore, the developmental arrest phenotype was rescued to the point where animals that contained a copy of the rescuing cosmid were able to grow to adult and produce progeny. Furthermore, most of the strains resulting from successful rescue experiments were maintained as viable stocks for at least two successive generations.
Physical map of the region of chromosome III deleted by the deficiencysDf127 and not covered by the duplication sDp8. The heavy horizontal line represents the chromosome. Dotted lines above the chromosome indicate sequenced cosmids that are not available as transgenic strains. All other cosmids represented above the chromosome are available as part of the transgenic library. Shaded boxes indicate cosmids used to rescue essential genes in this study. Gene names are written underneath the cosmids to which they have been correlated. An inverted V beneath the chromosome line indicates the primer sites used to determine the left end point of sDf127. A − or + under the inverted V indicates that the primer site is deleted or not deleted, respectively. The approximate region deleted by the deficiencysDf127 is shown as a single line below the chromosome. Regions of uncertainty are represented by a broken line. The approximate region covered by the left end of sDp8 is shown by a broken double line below the chromosome.
Essential Genes Correlated to Cosmids in This Study
DISCUSSION
We report here the construction of a Transgenic Library consisting of genetic strains carrying 250 cosmids “immortalized” as genetic duplications. Of these, 208 were constructed with purified cosmid DNA obtained directly from the C. elegans Genome Sequencing Consortium. To date, every cosmid injected has resulted in at least one heritable, stable line. In six cases, the concentration of cosmid DNA injected was lowered to generate transgenic strains successfully. In all cases, the final concentration of injected DNA was ∼100 ng/μl, made up by coinjected plasmid DNA containing therol-6(su1006) mutation and a kanamycin resistance gene to facilitate recombination with Lorist vectors (pCes1943). Heritable transgenics were identified initially by their semidominant Roller phenotype resulting from the coinjection. All strains were tested for the presence of vector sequences using PCR amplification with primers specific for the cosmid vectors. Our data showed that 72% of the strains constructed contained both the Rol-6 (pRF4) and cosmid vector sequences, and these were used to establish the Transgenic Library collection. For each of the cosmids injected, more than one strain was established whenever possible.
A subset of the transgenic strains has been tested for their ability to rescue visible and lethal mutations. Three visibles, which had been rescued previously, were tested and all were rescued by the predicted cosmids. In the case of the lethals, a reasonably small physical interval on chromosome III was defined by deficiency PCR. The interval between the left end of sDf127 and the left end ofsDp8 is covered by ∼15 sequenced cosmids, of which DNA for two cosmids was not available. Transgenic strains for the remaining 13 cosmids were crossed to lethal alleles representing the 10 identified essential genes that map to this region. Eight of the 10 genes were rescued by four of the cosmid transgenics. Maternal effect mutants exist for three of these genes, mel-32, let-725, andlet-721. Assuming successful progeny development is dependent on the expression of these genes in the germ line of the parent, it would seem, at least in the case of these three genes, that the extrachromosomal array responsible for rescuing the Mel phenotype is expressed in the germ line of transgenic animals. Therefore, it would appear that a broad range of phenotypes is rescuable by these transgenic strains.
Examination of the rescue data in a defined region of chromosome III illustrates how this project contributes to a systematic linkage of the genetic and sequenced physical map. A region of chromosome III was chosen for this analysis because of the availability of complete genomic sequence (Wilson et al. 1994). If we approximate the left endpoint of sDp8 to be the cosmid K03F8 (midway between cosmids C05D11 and M01G4), then the sDf127 interval contains just <100 coding elements defined by the program GENEFINDER (P. Green and L. Hillier, in prep.). Before this analysis none of these potential-coding elements were aligned to the genetic map. As a result of our analysis eight genetic loci have been assigned to cosmids. Thus, the map alignment in this interval has increased to an average resolution of 1 in 12 GENEFINDER-defined coding regions.
Increased map alignment facilitates gene cloning. The first step toward determining the position of a gene on the physical map is often to map the gene of interest to the genetic map between two cloned markers. This first step is made easier by increasing the number of genetic loci that have been positioned on the physical map. The next step is to estimate correctly the physical distance between the genetically mapped loci. A description of the problems associated with making these estimates for different genomic regions is presented in Barnes et al. (1995). The more accurate the “molecular/genetic” ruler for a particular region is, the more precise the prediction of physical location, and the more efficiently genes can be cloned and characterized. The availability of a collection of transgenic lines, each carrying a particular cosmid from the physical map, greatly streamlines the process of identification.
Annotated coding regions and computer-generated restriction maps have streamlined the procedure of correlating individual genes with mutant phenotypes. The ready availability of complete sequence data makes it possible to produce restriction maps in silico. It should be noted, however, that to avoid redundancy, the C. elegans Genome Sequencing Consortium reports only the unique sequence of each cosmid, plus enough overlapping sequence to confirm order, or keep a gene intact (Wilson et al. 1994). Thus, the DNA sequence reported in GenBank does not represent necessarily the actual size of the cosmid insert. When analyzing the sequence data of a cosmid that produces a positive rescue, entries that contain significantly more or less than 40–50 kb of sequence have most likely been edited. By analyzing the overlap information provided in the GenBank entries, the actual amount of cloned DNA can be calculated. This is necessary when identifying the actual coding region responsible for the rescue. However, if interpreted correctly, the sequence information provided by the sequencing consortium greatly streamlines the process of cutting down a rescuing cosmid and creating rescuing transgenics that carry single coding regions. This is currently being done for the cosmid C05D11 (G.P. Vatcher, unpubl.).
To assess the general usefulness of the transgenics, experimental tests of their rescuing abilities were done. Although our general method of PCR confirmation does not verify that the entire cosmid sequence is present in the extrachromosomal array, in all the cases examined, the rescue data indicate that intact cosmids are present. For the cosmid C05D11, rescues done using clones containing individual coding regions confirm rescues done using the complete cosmid (G.P. Vatcher, unpubl.). In this case, rescuing information demonstrates intact coding information for the cosmid transgenic. This is also at least partially indicated for the cosmid transgenics used to rescue the visible alleles of dpy-17, unc-32, and sma-3. The successful rescue of all three of these genes indicates that the majority of cosmid sequence is maintained in the extrachromosomal arrays through many generations. We have no evidence that the arrays degrade or break down over time.
We have no information about what types of genes might not be rescued by the transgenics. Two genes in the sDf127 region were not rescued, but we do not have complete transgenic coverage of this region. DNA for cosmids C18F10 and C56G2 was not available from the Genome Sequencing Consortium; therefore transgenics were not constructed immediately. The missing cosmids account for ∼11,000 bp of sequence and would be expected to contain two or three genes. Another possibility why these genes could not be rescued is that they are not fully contained on any one cosmid. Although it is not yet clear how often this can be expected to occur, it is predictable that some portion of genes will be unrescuable with a set of essentially nonoverlapping cosmids transgenics. Even when an entire gene appears to be contained on a cosmid, there is the possibility of upstream (or downstream) regulatory sequences that are not contained on the cosmid but are required for the correct expression of a gene.
We chose the Rol-6 marker for our constructions because it was widely used at the time the project began, and we have continued to use it. We realize that rescue of certain mutant phenotypes may be difficult to score in a roller background, but this will be a problem with most dominant markers. Despite the disadvantages, a dominant marker such asrol-6(su1006) is well suited for a large-scale project such as the transgenic library project because it allows the easy recovery and maintenance of the transgenic individuals. Using the Rol-6 marker has allowed the construction of transgenics in a wild-type background, and provided flexibility in transferring the arrays to a variety of other genetic backgrounds as required. Although we acknowledge that the transgenic strains we have created may not be useful for rescuing every gene, we have demonstrated that the transgenics can be generated, maintained, and genetically manipulated in large numbers. The ease of use, and potential to rescue large numbers of mutants, should make these transgenics a useful tool for the C. elegans research community.
The transgenics exist as genetic strains in which a cosmid is carried as a mini (unlinked) duplication. The extrachromosomal arrays can be transmitted between strains as for any unlinked duplication. The transgenic animals are healthy and easily mated as hermaphrodites. Even transgenic roller males can be used successfully in certain crosses, such as mating to Unc hermaphrodites. A single transgenic may be used repeatedly in different crosses to test for rescue of many different mutant phenotypes, or conversely a mutant strain may be tested against several different transgenics. Either method provides a rapid way to test for the rescue of one, or a number, of different mutant phenotypes.
As duplication strains, the transgenics have intrinsic value for genetic analysis, as some genes will be sensitive to dosage effects. One example of this was observed during our analysis. Two cosmid strains, overlapping on a specific region, produced males at a significantly higher frequency than expected. Availability of a complete transgenic collection makes possible comprehensive analysis of phenotypic effects caused by increased gene dosage.
Best estimates from quantitative PCR on these and other transgenic arrays indicates that the copy number of the cosmid in the array is in the range of one to five (R. Chiu, unpubl.). It is feasible that copy number or physical structure of extrachromosomal DNA may affect how genes contained in any particular array are expressed. In turn, this may have a dramatic effect on the ability of an extrachromosomal array to rescue mutations in some genes. Because we select transformed animals based on their exhibition of a strong Rol-6 phenotype, and because the severity of the Rol-6 phenotype is dependent on the copy number of the rol-6(su1006) gene (Kramer et al. 1990), we do not expect to isolate transformants that have an exceedingly low copy number of the rol-6 plasmid. However, we do expect that the copy number of cosmid DNA within the extra-chromosomal array may vary significantly between transgenic lines. Thus, multiple segregant lines were constructed for each cosmid whenever possible, and are available on request.
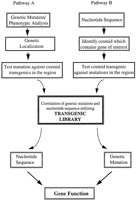
When the sequence of a putative C. elegans gene is known, it is possible to ask what phenotype a mutation in that gene will cause by screening for transposon insertions or subsequently induced deletions in the region of interest (Zwaal et al. 1993). When a genetic mutation is found that results in a particular phenotype, transformation/rescue experiments provide the C. elegans community with a complementary approach to correlating biological function with regions of DNA sequence (Fig. 4). With clean, complete sequence data readily accessible, potential coding regions can be identified quickly and sequence restriction maps generated in silico. An accessible cosmid transgenic library, such as the one we are constructing, eliminates the need for individual laboratories to construct large numbers of transgenic strains for gene mapping purposes. Instead, researchers can focus their microinjection experiments on determining which gene contained within a cosmid (or cosmids) rescues the mutation of interest. Once a gene has been isolated, a vast array of molecular techniques, such as expression analysis, two-hybrid protein interaction analysis, and site-directed mutagenesis can be brought to bear on the determination of function. It is this kind of analysis that is required to gain a true understanding of how the genomic sequence of an organism is processed to create a living organism.
An overview of how the cosmid transgenic library constructed in this study can be used in the process of determining the biological function of a nucleotide sequence. If a mutant phenotype has been assigned to a particular gene, pathway A can be used to determine what the nucleotide sequence of that gene is. If a putative gene has been identified based on sequence similarity, pathway B can be used to isolate genetic mutations in the gene of interest.
METHODS
The nomenclature used conforms to that proposed by Horvitz et al. (1979).
Nematode Strains and Culture Conditions
All strains were maintained and mated on Petri plates containing nematode growth medium (NGM) streaked with Escherichia colistrain OP50 (Brenner 1974). All experiments were carried out at 20°C (Rose and Baillie 1979). The wild-type strain N2 (Bristol) used to create the transgenic strains is a derivative of an N2 strain received from MRC, Cambridge, UK.
Lethal Mutations sDp8 and sDf127
All lethal mutations used in this study are recessive and, except for mel-32, were generated in ethylmethane sulfonate (EMS) mutagenesis screens (H.I. Stewart, in prep.). Alleles ofmel-32 were generated by G.P. Vatcher in an EMS mutagenesis screen for maternal effect lethals. We define a lethal mutation (let) as any mutation that prevents an organism from producing viable progeny. This includes mutations that result in the developmental arrest of the animal, and those that allow the animal to grow to adult, but are sterile. Mutations that have no visible effect on the homozygous animal itself, but result in progeny exhibiting a lethal phenotype are referred to as maternal effect lethals (mel). The duplication sDp8(s1957)(III;I) is described by Stewart et al. (1991). The deficiencysDf127(s2428)(III) was generated in a UV mutagenesis screen for rearrangements that result in lethal phenotypes (H.I. Stewart, in prep.).
Cosmid Injection
Cosmid DNA used to create transgenic worms was obtained in one of two ways. For as many cosmids as possible, purified DNA was received directly from the C. elegans sequencing laboratories in St. Louis, MO, and Hinxton, UK. However, the sequencing laboratories were unable to provide DNA for all cosmids. For the missing cosmids, bacterial clones containing sequenced cosmids were received from Alan Coulson (of the C. elegans sequencing laboratory in MRC, Hinxton, UK). DNA was prepared using the Pharmacia Mini-Prep Kit Plus (minus the final column purification step). Cosmid DNA was diluted in TE (10 mm Tris-HCl at pH 8.0; 1 mm EDTA at pH 8.0) and mixed with pCes1943 (S. Jones and B. Barbazuk, unpubl.), a plasmid derived from pRF4 (Kramer et al. 1990). pCes1943 contains the semidominant rol-6(su1006) allele (Mello et al. 1991), and has been modified to contain the kanamycin resistance gene. Concentrations that we report for our injection experiments are only estimates, based on either gel quantification or information provided to us by the Sequencing Consortium. The final concentration of injection mixtures used for the majority of cosmids was ∼80 ng/μl pCes1943 + ∼20 ng/μl cosmid DNA. The concentration of 20 ng/μl was decreased for cosmids that did not result in stable transformants (see Table 2), or for cosmids for which limited amounts of DNA were available. Before injection, the DNA mixture was centrifuged for 20 min to settle any particulate matter. Injections were performed on wild-type hermaphrodite animals using the methods described by Mello et al. (1991). After injection, worms were placed individually onto plates and resultant progeny were screened for the presence of worms exhibiting the Rol-6 phenotype. These F1animals were picked individually to fresh plates and allowed to self-fertilize. The F2 generation was again screened for Rol-6 worms and lines that continued to segregate roller progeny were tested, using PCR, for the presence of cosmid vector (see below). Worms that tested positive for the presence of vector were assumed to be carrying an extrachromosomal array containing both pCes1943 and the desired cosmid DNA. Extrachromosomal arrays were assigned asEx number (N) (s being our laboratory designator; Ex meaning extrachromosomal array), and the strains carrying them were preserved cryogenically for future use as +/+; sExN [cosmid + pCes1943].
Testing Transgenic Strains by PCR
Template DNA was extracted from transgenic worms exhibiting the Rol-6 phenotype as per Barstead et al. (1991) with modifications ofWilliams et al. (1992). One or two worms were digested for each 25-μl PCR. Thermocycling reactions were performed in an Idaho Technology model 1605 Air Thermo-cycler using Low [20 mmMg2+] 10× reaction buffer obtained from Idaho Technologies. Taq polymerase was obtained from BioCan Scientific. Nucleotide triphosphates were obtained from Pharmacia. Thermal profiles for all PCR reactions were 1 min at 94°C, followed by 30 cycles of 10 sec at 94°C, 20 sec at 59°C, 40 sec at 72°C, followed by a final extension of 2 min at 72°C. Ramp speed was set at 9 (fastest).
Oligonucleotide primers specific for Lorist cosmid vector sequences, Krp17 (5′-CGTCCGGCGCACAGAAGC-3′) and Krp18 (5′-GTGCTGAGCCCGGCCAAA-3′), were designed by S. McKay (1993). Primers DC1 (5′-GCGGGGTTGCCTTACTGG-3′) and DC2 (5′-CGCAAGGTCGGGCTGAAC-3′) are specific for pJB8 cosmid vector sequences. Positive control primers R5 (5′-TGATGATGGATTGGCTCGGC-3′) and R11 (5′-TACTCGCATCTTTACCATCG-3′) were designed by M.A. Marra (Genome Sequencing Center, Washington University School of Medicine, St. Louis, MO) and are specific for chromosome IV sequences.
The control primers R5 and R11 were included in every reaction. This allowed us to differentiate between reactions that failed to give an experimental band because the template worm did not contain cosmid DNA, from reactions that did not work for other technical reasons. Test reactions with both control and experimental primers revealed minimal primer interaction. The expected product size of the control primers is 380 bp, whereas the product sizes of the Lorist- and pJB8-specific primers are 880 and 618 bp, respectively. These bands are easily distinguished on a 1% agarose gel (see Fig. 2).
Deficiency Mapping Using PCR
When homozygous, the deficiency sDf127 results in animals that exhibit embryonic lethality. Fifteen hermaphrodites of the genotype sDp3(III;f); [dpy-17(e164) sDf127(s2428)] unc-32(e189) III (see below) were transferred to a seeded plate and allowed to lay eggs for 8 hr. Adult hermaphrodites were removed and the plates were kept at 20°C for 48 hr to allow viable eggs (containingsDp3) to hatch. Eggs that fail to hatch after this time were considered to be of the genotype dpy-17 sDf127 unc-32. For each reaction, one or two arrested eggs were treated with chitinase and transferred to worm lysis buffer (see above). PCR reactions were performed as above with the annealing temperature being modified to suit the primers used. Primers were designed based on cosmid sequences available from GenBank. All PCR reactions were run in duplicate with positive control primers. Cosmid primers that produced no PCR product indicate that the cosmid sequence from which at least one of the primers was designed was deleted by sDf127 (assuming the control band was present). All such negative results were retested to confirm results. Cosmids tested in this way are shown in Figure 3.
Rescue of the Visible Mutations dpy-17(e164), sma-3(e491), and unc-32(e189)
The following double mutants were used to test visible mutations for rescue: dpy-17(e164) unc-36(e251) (to testdpy-17), sma-3(e491) unc-36(e251) (to testsma-3), and dpy-17(e164) unc-32(e189) (to testunc-32). Transgenic strains containing the appropriate cosmid were crossed with N2 males. Resulting Rol-6 males of the general genotype +/+;sExN were mated with homozygous, double mutant hermaphrodites as shown above. From the progeny of this cross, L4 Rol-6 hermaphrodites were set individually to plates and allowed to self-fertilize. The resulting progeny were screened for animals that gave Unc-36 (or Dpy-17 in the case of the unc-32 cross) progeny that did not express the phenotype of the visible mutation being tested for. These were set individually to plates and allowed to produce progeny to verify that the visible mutation being tested was still present.
Rescue of Recessive Lethal Mutations Using Transgenic Strains
Lethal (let or mel) Mutations
Mutant strains have the general genotype sDp3(III;f); dpy-17(e164) let-x(sy) unc-32(e189) III. The let ormel mutations are recessive and are complemented by the free duplication sDp3 that covers a large portion of chromosome III (left), including dpy-17. The right break point ofsDp3 is between unc-36 and unc-32 [as per ACeDB (A C. elegans Database; R. Durbin and J. Thierry Mieg 1991–). Documentation, code, and data available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk, andncbi.nlm.nih.gov]; therefore, homozygous let ormel-bearing strains that carry sDp3 exhibit a Unc-32 phenotype. The let and mel mutations were generated using EMS, and mapped to the right of dpy-17 (H. Stewart, in prep.).
Lethal (let or mel) Rescue
The general complementation test procedure that we used to test for cosmid rescue of lethal mutations is as follows. Males heterozygous for a lethal mutation and having the general genotype (sDp3); dpy-17(e164) let-x(sy) unc-32(e189)/+++, were constructed by crossing lethal-bearing hermaphrodites to wild-type males. Although some heterozygous males produced from this cross will carry the duplication, its presence was not selected for and does not effect the qualitative outcome of the test. Therefore, for the sake of simplicity, it will be assumed lost in subsequent genotypes. Transgenic hermaphrodites containing the cosmid of interest were mated with heterozygous lethal-bearing males for 36 hr. Individual hermaphrodites were then removed to separate plates and allowed to produce progeny. The F1 progeny were screened for Rol-6 males, which indicated that outcrossing had occurred. From plates that contained outcrossed progeny, 25-40 L4, Rol-6 hermaphrodites were picked individually to fresh plates and allowed to self-fertilize. These individuals were of two possible genotypes dpy-17(e164) let-x(sy) unc-32(e189)/+++; sExN or +/+;sExN. The progeny from these animals were screened for the presence of arrested or dying Dpy-17 Unc-32 animals. The presence of these animals indicated that the lethal mutation of interest was present in the F1 hermaphrodite. Plates that contained arrested animals were then screened for the presence of Dpy-17 Unc-32 animals that failed to arrest (the Rol-6 phenotype is masked by the Unc-32 phenotype). A minimum of five such plates containing a minimum of 30% roller progeny were screened before a negative rescue result was considered. If present, adult Dpy-17 Unc-32 individuals were removed to fresh plates and tested for fertility. The existence of fertile animals suggested that the lethal allele was rescued by the extrachromosomal array. In the case of maternal effect lethals, the fertility test was continued for another generation. If thesDp3 duplication was present in the F1hermaphrodite, then the number of Dpy Unc animals would be reduced. However, any animals that exhibit the definitive Dpy Unc phenotype cannot carry the duplication, as sDp3 covers dpy-17.
In each case, positive rescue results were obtained using our canonical transgenic strain. In some cases, the rescue result was confirmed using a different segregant, but no variance in results was observed.
Confirming the Rescue
Putative rescued animals were tested in the following ways.
PCR ANALYSIS
Between 8 and 10 fertile animals, putatively of the genotypedpy-17 let-x unc-32; sExN were used as a template to confirm the presence of cosmid DNA (see Testing Transgenics strains by PCR). As a negative control, developmentally arrested worms were used. In this manner the rescue phenotype was correlated with the presence of cosmid DNA.
OUTCROSS ANALYSIS
Fertile animals putatively of the genotype dpy-17 let-x unc-32; sExN were crossed to wild-type males. The progeny of this cross were scored for the presence of Rol-6 animals (indicating the presence of the extrachromosomal array). From these progeny, 25 L4 hermaphrodites that did not exhibit the Rol-6 phenotype (putatively of the genotype dpy-17 let-x unc-32/+++) were placed individually on plates and allowed to produce progeny. These progeny are then screened for the presence of Dpy-17 Unc-32 animals that displayed the terminal phenotype normally associated with the lethal mutation. If all Dpy Unc progeny arrest, the continued presence of the lethal mutation was confirmed.
REPEAT CROSS
All rescue experiments were performed at least twice for confirmation.
All successfully rescued lethal mutations were tested against at least one nonrescuing cosmid transgenic to eliminate nonspecific rescue activity.
Acknowledgments
This work is being done in collaboration with the C. elegans genome sequencing laboratories. We thank Dr. Bob Waterston and members of the Genome Sequencing Center in St. Louis, MO, and John Sulston and members of the Sanger Centre in Hinxton, UK, for providing sequenced cosmid DNA for this project. A special thank you goes to M. Marra and R. Wilson (St. Louis) and S. Jones and A. Coulson (Hinxton) for putting up with our never-ending cosmid requests. Also, thanks to E. Gilchrist, R. Rosenbluth, and H. Janke for their editorial comments and support, and to the C. elegans community on a whole, whose support and feedback was essential to making this project work. The Transgenic Library Project was funded by a grant from the Canadian Genome Analysis and Technologies Program (CGAT) to A. Rose and D. Baillie. The deficiencies and lethal mutations were generated by The Genetic Toolkit project, which is funded by a grant from the National Institutes of Health National Center for Research Resources (NCCR) (USA) to A.R., D.B., and D.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL djanke{at}darwin.mbb.sfu.ca; FAX (604) 291-4597.
-
- Received June 30, 1997.
- Accepted August 8, 1997.
- Cold Spring Harbor Laboratory Press