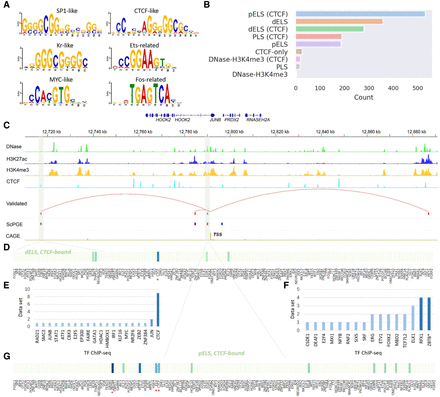
ScPGE discovers important motifs in specific types of cCREs. (A) Some important TF motifs found in true positives categorized by ScPGE, which are labeled by TF family names. (B) The distribution of types of cCREs, sourced from the SCREEN Registry V3, where pELS means proximal enhancer-like signatures, dELS means distal enhancer-like signatures, and PLS means promoter-like signatures. (C) Visualization of active cCREs of the JUNB gene identified by ScPGE, where validated cCREs are represented by red rectangles and correctly identified cCREs are represented by blue rectangles with red outlines. (D,E) The contributions of TF motifs in a type-specific cCRE (dELS, CTCF-bound), where significant TFs are supported by TF ChIP-seq data sets. (F,G) The contributions of TF motifs in a type-specific cCRE (pELS, CTCF-bound), where significant TFs are supported by TF ChIP-seq data sets.











