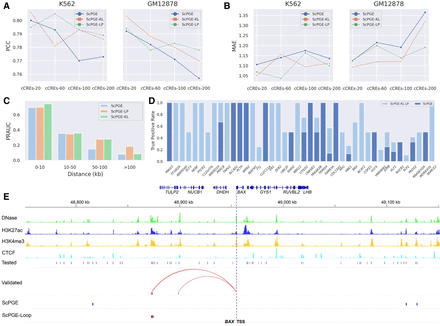
Chromatin loops facilitate ScPGE to capture cCRE–gene interactions. (A,B) Comparison of the performance of ScPGE, ScPGE-KL, and ScPGE-LP as the number of cCREs increases. (C) The performance (PRAUC) of ScPGE, ScPGE-KL, and ScPGE-LP in classifying cCRE–gene pairs at different distance groups. (D) The true positive rates of ScPGE and ScPGE-Loop on validated ccREs. (E) Visualization of active cCREs of the BAX gene missed by ScPGE but identified by ScPGE-Loop, where validated cCREs are represented by red rectangles and correctly identified cCREs are represented by blue rectangles with red outlines.











