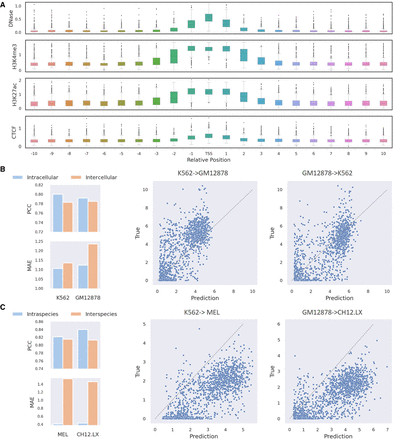
ScPGE discovers different patterns by analyzing predictions. (A) A pattern found in true positives (TPs) that the regulatory effect of cCREs on target genes decreases with distance. (B) The cross-cell-type predictive performance of ScPGE, where K562→GM12878 indicates using a model trained on K562 data to predict GM12878 data. (C) The cross-species predictive performance of ScPGE, where K562→MEL indicates using a model trained on K562 data (Human) to predict MEL data (Mouse).











