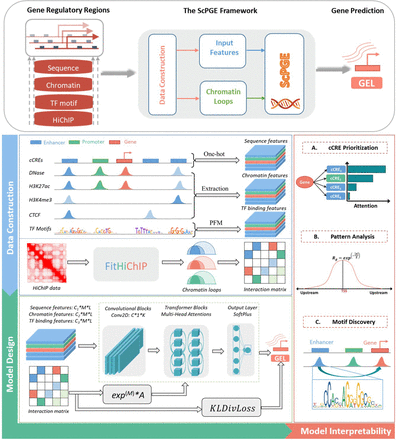
The framework of ScPGE. The framework predicts gene expression by integrating sequence features, chromatin features, TF motif features, and HiChIP interactions of discrete candidate cCREs on both sides of genes. During the stage of data construction, we transformed sequence features, chromatin features, and TF motif features into three-dimensional tensors and converted chromatin loops identified by FitHiChIP into interaction matrices. During the stage of model design, we combined CNN and transformer to predict gene expression, where CNN is used to learn sequence and chromatin features and transformer is used to learn the relationships between genes and cCREs. Meanwhile, we designed two ways to improve the performance of ScPGE by incorporating chromatin loops. In the stage of model interpretability, we performed cCRE prioritization, pattern analysis, and motif discovery.











