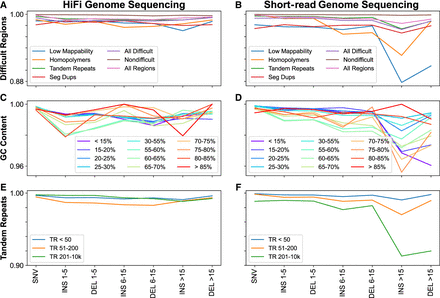
SNV/indel accuracy across GIAB-defined genomic regions. Small variant F1-scores for HiFi genome sequencing (A,C,E) and short-read genome sequencing (B,D,F) across 22 of the 100 interrogated GIAB-defined genomic regions. Results summarized by difficulty (mappability, homopolymers, tandem repeats, segmental duplications, all difficult, not in any difficult, all regions), GC content (<15% to >85%), and tandem repeats (<50 bp, 51–200 bp, 201–10,000 bp), and stratified by variant type (SNV, indels 1–5 bp, 6–15 bp, and ≥16 bp).











