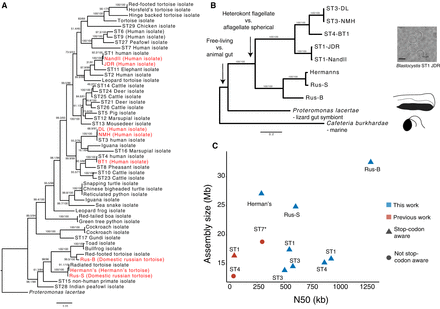
Phylogenetic relationships and genome assembly characteristics of Blastocystis. (A) 18S rRNA maximum likelihood (ML) phylogeny of Blastocystis subtypes, with Proteromonas lacertae as an outgroup. Support values are 1000 replicates of ultrafast bootstrap and SH-aLRT tests (Guindon et al. 2010; Hoang et al. 2018). Strains in red were sequenced in this study. (B) 13-gene concatenation ML phylogeny for all strains and species with sequenced genomes used in this study. Important evolutionary transitions are highlighted. Images represent cellular morphology of Cafeteria burkhardae, Proteromonas lacertae, and Blastocystis. (C) Assembly statistics of Blastocystis strains sequenced in this work compared with previously sequenced and annotated genomes. Stop-codon aware and not stop codon aware designations refer to gene annotation methodologies that take into consideration that Blastocystis genes can lack stop codons or do not consider this, respectively. The ST7 N50 is a gapped scaffold N50 generated with Sanger sequencing scaffolding.











