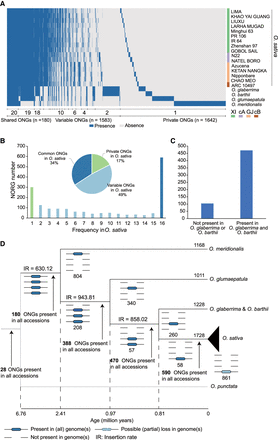
Abundant presence/absence variations of ONGs in Oryza genomes. (A) Presence and absence information of ONGs in five AA genome species. The presence of a NORG is indicated by blue, and the absence of a NORG is indicated by white. (B) Distribution of ONGs among 16 O. sativa accessions. The “common ONGs” are those present in all accessions; “variable ONGs” are present in more than one but fewer than 16 accessions; “private ONGs” are present in only one accession. (C) Distribution of O. sativa “common ONGs” in O. glaberrima and O. barthii. (D) Position of ONGs on the phylogenetic tree of AA genome species in Oryza. The total number of ONGs in genomes is shown above branches. The number of branch-specific ONGs is shown below branches. The four lines under each node from top to bottom represent O. meridionalis, O. glumaepatula, O. glaberrima and O. barthii, and O. sativa.











