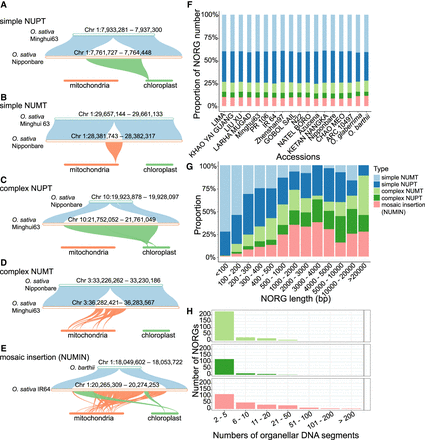
NORG types vary depending on length but are consistent across Oryza genomes. (A–E) Structure of five types of NORGs. (A) A simple NUPT consisting of one segment from plastid. Synteny between Minghui63 Chr 1: 7,933,281–7,937,300, ONG0100220 in Nipponbare, and the plastid of rice. (B) A simple NUMT consisting of one segment from mitochondria. Synteny between Minghui63 Chr 1: 29,657,144–29,661,133, ONG0100760 in Nipponbare, and the mitochondria of rice. (C) A complex NUPT consisting of two segments from the plastid. Synteny between Nipponbare Chr 10: 19,923,878–19,928,097, ONG1000620 in Minghui63, and the plastid of rice. (D) A complex NUMT consisting of eight segments from the rice mitochondria. Synteny between Nipponbare Chr 3: 33,226,262–33,230,186, ONG0301080 in Minghui63, and the mitochondria of rice. (E) A nuclear complex insertion (NUMIN) consisting of two segments from plastid and 26 segments from the mitochondria. Synteny comparison between O. barthii Chr 1: 18,049,602–18,053,722, ONG0100500 in O. sativa IR64, and the mitochondria and the plastid of rice. (F) Relative proportion of five types of NORGs in each accession. (G) Relative proportion of each type of NORGs with different lengths. The length of NORGs was calculated using the median size. (H) Number of organelle DNA segments in complex NORGs.











