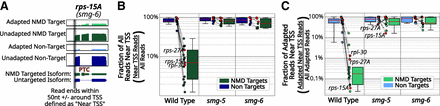
NMD-targeted transcripts exhibit low levels of decapped RNAs in wild-type animals. (A) Diagram illustrating how near trans-splicing site (TSS) 5′ ends are defined. As in Figure 3B, the coverage plots show read pileups on the gene rps-15A. Reads with their 5′ ends within 50 nucleotides of annotated TSSs are labeled as “near TSS.” This process was carried out for NMD-target and nontarget isoforms. (B) Box and strip plots of the fraction of all reads with 5′ ends spanning to the TSS for each isoform. Red solid lines indicate the median. The inner box denotes the inner quartile range (IQR) from the 25th to the 75th percentiles; whiskers span out to any points within 1.5 times the IQR. Each point indicates the mean for each isoform across replicates. Within each strain (x-axis), reads were separated based on isoform identity as non-NMD-targets (left; blue) or NMD-targets (right; green). Example genes shown in other figures are indicated by red points (rpl-30, rps-15A, and rps-27A) and labeled where not tightly clustered. Only isoforms that contained at least one adapted read in at least two replicates were considered. Dashed lines between points connect isoforms within strains from the same gene loci. Box and strip plots (as in B) of the fraction of all adapted reads near the TSS.











