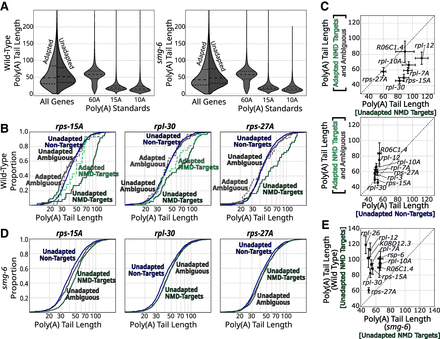
NMD-target mRNAs and their degradation products have poly(A) tails that are as least as long as normal mRNAs. (A) Violin plots for all mRNAs in wild-type and smg-6 animals. The left side of each violin (in light gray) shows the distribution of adapted reads’ tail lengths, and the right (in dark gray) shows unadapted reads. The three violins to the right show the tail length distributions for three spike-in RNA standards. Long dashed lines indicate the means, and short dashed lines indicate first and fourth quartile boundaries. (B) Poly(A) tail length cumulative distribution function (CDF) plots of example genes (rps-15A, rpl-30, and rps-27A) in wild-type animals. The same color scheme is used here as in Figure 3: adapted NMD isoforms (dashed light green), unadapted NMD isoforms (dark green), adapted non-NMD isoforms (dashed light blue), unadapted non-NMD isoforms (dark blue), adapted ambiguous isoforms (dashed light gray), and unadapted ambiguous isoforms (dark gray). For each plot, only categories that had at least 10 poly(A) tail-called reads are shown. For statistical analysis, see also Supplemental Table S4. (C) Scatter plot comparisons of mean poly(A) tail lengths between adapted versus unadapted NMD targets and adapted NMD-target reads versus unadapted nontargets for genes in which we could identify the NMD-target isoform. The dashed line indicates the diagonal where X = Y. Error bars indicate the SEM. Only genes with at least 10 reads in each category are shown. (D) Poly(A) tail length CDF plots of example genes in smg-6 animals (as in B). Low read counts caused adapted, PTC-mapping RNA species to fall below the 10 read cutoff. For statistical analysis, see also Supplemental Table S4. (E) Scatter plot comparison of poly(A) tail lengths between unadapted NMD targets in wild-type animals versus smg-6 animals.











