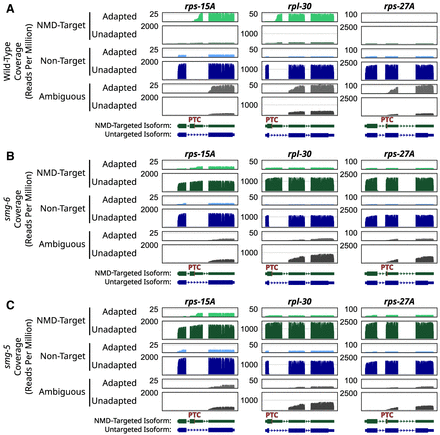
Nanopore degradome sequencing captures smg-6- and smg-5-dependent degradation intermediates on NMD targets. (A) Coverage plots of loci (rps-15A, rpl-30, and rps-27A) in wild-type animals that produce NMD-target and nontarget mRNAs. Read coverage (y-axes) is shown in reads per million. From top to bottom, coverages are for the following categories: adapted NMD isoforms (light green), unadapted NMD isoforms (dark green), adapted non-NMD isoforms (light blue), unadapted non-NMD isoforms (dark blue), adapted ambiguous isoforms (light gray), and unadapted ambiguous isoforms (dark gray). Annotations at the bottom indicate the NMD-targeted isoform and the non-NMD-targeted isoform. (PTC) Location of the NMD-eliciting stop codon. (B) Coverage plots of the indicated loci in smg-6-mutant animals. (C) Coverage plots of the indicated loci in smg-5-mutant animals.











