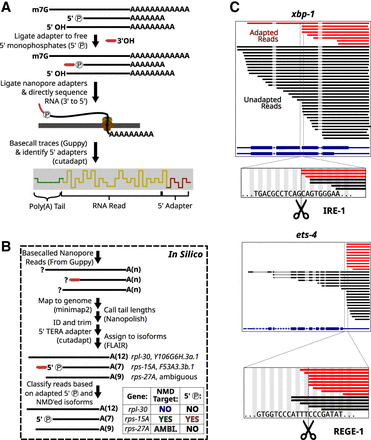
Workflow for the capture of mRNA degradation intermediates. (A) Strategy to identify RNAs with a 5′ monophosphate using direct RNA sequencing of ONT. For details, see Methods. (B) In silico read processing workflow for degradome sequencing. For details, see Methods. (C) RNAs with 5′ monophosphate ends produced from known endonuclease targets xbp-1 and ets-4. (Top) Red lines indicate molecules that contained the 5′ adapter sequence and were thus derived from 5′ monophosphate–containing RNAs. Black lines indicate molecules that did not contain the 5′ adapter sequence. Thick lines indicate aligned sections of the RNA sequences, and thin lines indicate alignment gaps of RNA reads, which span annotated introns. The isoform annotations for xbp-1 (and ets-4) are indicated in blue. The thickest sections of the isoform annotations indicate the coding sequences. (Bottom) A zoomed-in window showing reads with 5′ ends near the known endonuclease cleavage sequence and site (indicated with dotted line and scissors).











