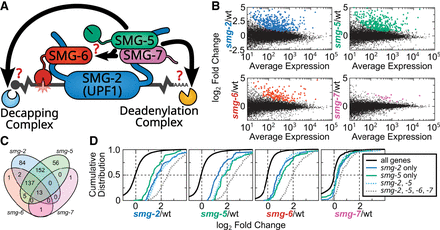
C. elegans UPF1, SMG-5, and SMG-6 repress a common set of endogenous mRNAs. (A) Model of decay step(s) of NMD on an mRNA target. (B) Differential expression analysis (DESeq2) of biological replicates of RNA-seq from wild type, smg-2 (UPF1), smg-5, smg-6, and smg-7. Genes with mRNAs that are upregulated in the indicated mutant (adjusted P ≤ 0.01) are colored. (C) A four-way Venn diagram indicating overlap of genes with mRNAs upregulated in the indicated mutants. The slight difference in numbers of targets compared with A (e.g., 16 vs. 15 smg-7 targets) is caused by taking the intersection of all genes included by DESeq2 across all four smg mutants. (D) Expression of all genes’ mRNAs (black) across smg mutants and grouped by whether the genes’ mRNAs are upregulated in smg-2 only (blue line; 84 genes), smg-5 only (green line; 56 genes), smg-2 and smg-5 (light blue dotted line; 152 genes), or smg-2, smg-5, and smg-6 and/or smg-7 (gray dotted line; 150 genes). All changes are statistically significant relative to all genes (KS test P <1 × 10−5).











