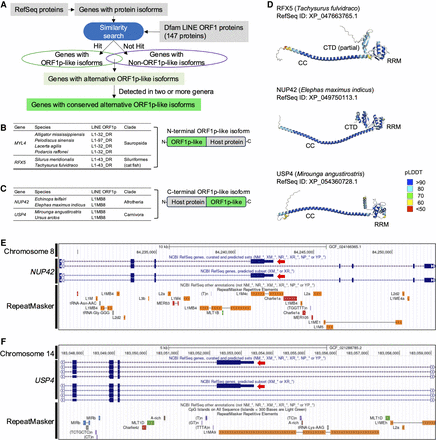
Other ORF1p fusion isoforms in vertebrates. (A) Schematic workflow of the detection of ORF1p fusion isoforms from RefSeq proteins. (B) In the nonmammalian vertebrate RefSeq proteins, two protein-coding genes had alternative ORF1p-like exons. The top hit LINE ORF1p was described in each fusion isoform. In both genes, the amino acid sequences encoded by the first coding exons were similar to ORF1p. (C) In the mammalian vertebrate RefSeq proteins, two protein-coding genes had alternative ORF1p-like exons. The amino acid sequences encoded by the final coding exons were similar to ORF1p. (D) Protein structures of the ORF1p-like amino acid sequence encoded by the noncanonical exons predicted by AlphaFold2. Coiled-coil (CC), RNA recognition motif (RRM), and C-terminal domain (CTD). (E,F) UCSC Genome Browser views of ORF1p-like isoform with RepeatMasker tracks. Red arrows indicate the ORF1p-like exons. The UCSC Genome Browser was accessed on June 25, 2024. NUP42 in the Elephas maximus indicus (Indian elephant) genome assembly (GCF_024166365.1; E), and USP4 in the Mirounga angustirostris (Northern elephant seal) genome assembly (GCF_021288785.2; F).











