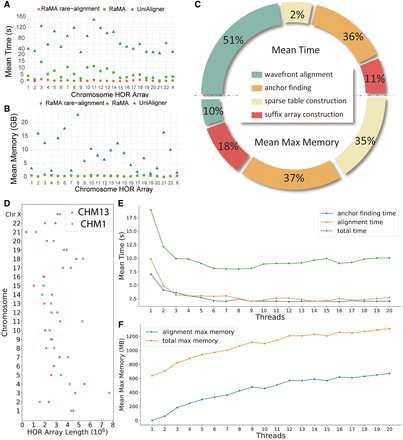
Comprehensive performance evaluation of RaMA: time and memory efficiency metrics. (A) Comparison of the average alignment time for HOR arrays across different chromosomes of CHM13 and CHM1 using RaMA and UniAligner. (B) Comparison of the average maximum memory usage for HOR arrays across different chromosomes of CHM13 and CHM1 using RaMA and UniAligner. (C) Comparison of the time and memory required for different algorithm stages of RaMA. (D) Comparison of HOR array lengths across different chromosomes between CHM13 and CHM1. (E) Variation in RaMA's anchor finding time, alignment time, and total time with the number of threads. (F) Variation in RaMA's alignment memory and total memory usage with the number of threads.











