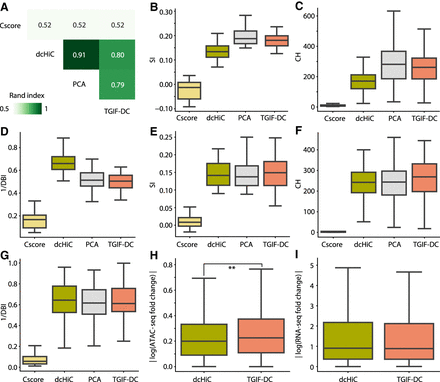
Benchmarking TGIF-DC on data from H1 and H1 differentiated to definitive endoderm. (A) Similarity of compartment assignments from different methods measured by Rand index. (B–D) Quality of compartments based on O/E counts measured by the silhouette index (SI; B), Calinski–Harabasz score (CH; C), and Davies–Bouldin index (DBI; D). (E–G) SI (E), CH (F), and DBI (G) on accessibility (ATAC-seq) signal. (H) Magnitude of log fold change in accessibility between H1 and endoderm within sigDCs identified by dcHiC and TGIF-DC. (I) Magnitude of log fold change in gene expression between H1 and endoderm within sigDCs identified by dcHiC and TGIF-DC.











