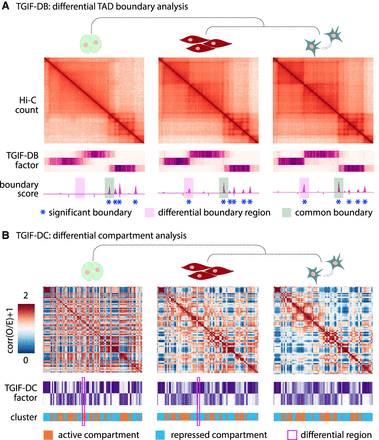
Overview of TGIF. (A) TGIF for differential boundary analysis (TGIF-DB). TGIF-DB takes multiple Hi-C count matrices as input and simultaneously learns a lower-dimensional representation of genomic regions based on their interaction patterns. The input matrices are from related biological conditions with their relationship encoded as a tree. From the lower-dimensional factors, we measure the boundary score of each region and identify boundaries for each input condition and significantly differential boundaries (sigDBs) for every pair of conditions. (B) TGIF for differential compartment analysis (TGIF-DC). TGIF-DC converts input matrices into correlation matrices of observed-over-expected (O/E) counts and factorizes them to yield latent features, which are used to cluster the regions. Each cluster corresponds to a compartment or a subcompartment. TGIF-DC also identifies significantly differential compartmental regions (sigDCs) for every pair of input conditions.











