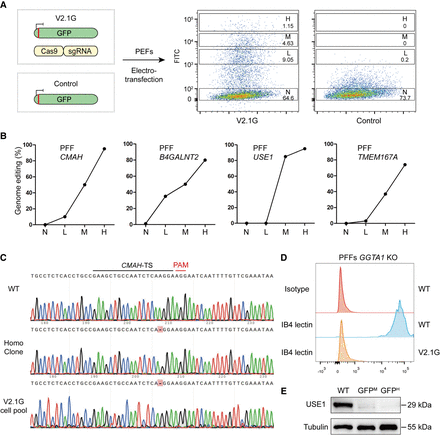
Selection of homozygously edited cells based on highest surrogate reporter expression. (A) Schematic of selection strategy using version 2.1-eGFP (V2.1G), a nonintegrated surrogate reporter system. PFFs were cotransfected with V2.1G, Cas9, and sgRNA plasmid, whereas the control group was only transfected with the V2.1G plasmid. FACS analysis revealed varying intensities of FITC expression among the cells. (H) The highest FITC intensity population, (M) the middle FITC intensity population, (L) the lower FITC intensity population, and (N) the negative FITC intensity population. (B) Correlation between genome editing efficiency and FITC intensity of endogenous targeting in PFFs. (C) Representative Sanger sequences of CMAH target site in wild-type cells, homozygous cell clones, and cell pools selected by V2.1G. (D) FACS analysis of GGTA1 targeting in PFF pools detection by IB4 lectin staining. (E) Western blot analysis of USE1-targeted Vero E6 cell pools with middle and high FITC intensity, respectively.











