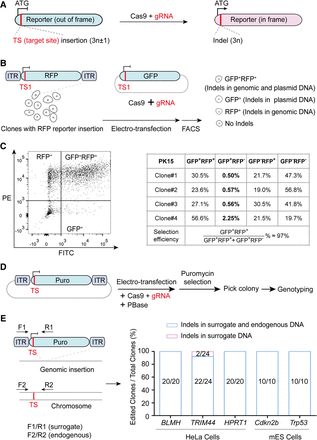
Detection of targeting events using surrogate reporters. (A) The principle of the surrogate reporter system. The reporter gene is initially frameshifted owing to the insertion of a CRISPR target site adjacent to the “ATG” start codon. CRISPR-Cas9 mediated editing results in insertions and deletions (indels) that restore the correct reading frame of the reporter gene. (B) Workflow for assessing the correlation between genomic and plasmid DNA targeting at the single-cell level. Colonies containing an integrated surrogate reporter (eRFP) were electro-transfected with Cas9, sgRNA, and a surrogate reporter (eGFP) plasmid. Both genomic (eRFP) and plasmid (eGFP) surrogate reporters incorporate the same target site (TS1). (ITR) Inverted terminal repeat sequence. (C) Flow cytometry analysis distinguishing colonies edited in genomic DNA (PE positive) and plasmid DNA (FITC positive). The selection efficiency is calculated by the ratio of GFP and RFP double-positive cells to GFP-positive cells. (D) Workflow for assessing the correlation between endogenous and surrogate DNA targeting at the single-cell clone level. Cells were electro-transfected with Cas9, sgRNAs, a surrogate reporter (Puro), and PBase. Following puromycin selection, cell colonies were picked for genotyping. (E) Primers (F1/R1) amplify the surrogate target site (Puro) integrated into the genome, and primers (F2/R2) amplify the endogenous locus. The right panel illustrates the ratio of edited clones.











