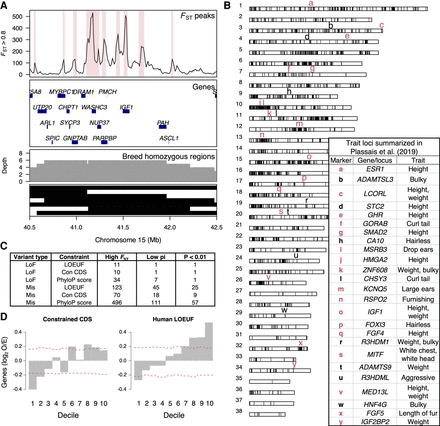
Noncoding variation at constrained genes drives breed differentiation. (A) Identification of breed-differentiated regions at Chromosome 15 IGF1 locus. Genomic windows with FST > 0.8 between breed pairs were plotted across the genome. Peak FST regions were then classified as regions of breed differentiation. Top FST peaks are shown in the first panel and highlighted in red. The second panel shows genes found in the region. The third panel shows the density of regions of breed homozygosity that were identified as 1 Mb genomic regions with breed nucleotide diversity <0.02. The final panel shows black bars as individual breed homozygosity stretches. (B) Genome-wide distribution of breed-differentiated regions. Trait loci summarized in Plassais et al. (2019) are displayed as lower-case characters. Trait loci in red are within 100 kb of a breed-differentiated region. (C) Number of genes in highly differentiated regions that contain potentially functional variants that pass minimum constraint thresholds. The “High FST” column is the number of genes that contain variants of interest. The “Low pi” column is the number of genes that contain variants in breeds with low nucleotide diversity across breed-differentiated regions. The final column is the number of genes that contain variants associated with breed differentiation (Fisher's exact test, P-value < 0.01). (D) Observed over expected frequency of constrained genes within breed-differentiated regions. The x-axis indicates constraint decile with 10 being the most constrained genes.











