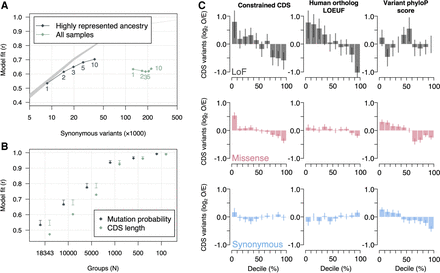
Rare canine variants are used to estimate mutational tolerance in constrained genes. (A) Pearson correlation coefficient between observed rare variants and gene mutation probabilities. Variants are organized by the group of samples they were observed in and the maximum allele count threshold for defining rare variants, which is displayed near each data point. The gray band represents the 95% expected interval of the model fit based on simulated accumulation of synonymous mutations. (B) Pearson correlation coefficient of rare variant frequency and gene mutation probability across gene groups. Genes were grouped according to their mutation probabilities. The x-axis shows the number of groups used to calculate correlation coefficients. The number of groups decreases as group sizes get larger. Two models are tested; one is the mutation probabilities calculated using the mutation model and the other is mutation probabilities according to gene CDS length. The 95% confidence intervals for the expected model fit are also displayed. (C) Gene mutation frequency according to three different constraint metrics. The expected number of mutations was calculated following the mutation model. The observed number of mutations are from singletons in dogs with highly represented ancestry. Error bars indicate whether the observed mutation frequency is outside the 95% expected bounds.











