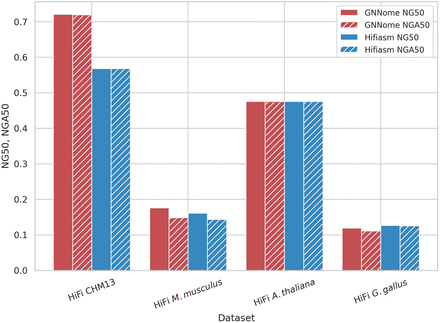
Contiguity on different haploid data sets. Comparison of contiguity of assemblies generated with GNNome and hifiasm (Cheng et al. 2021) for HiFi data. The NG50 of the assembly is defined as the sequence length of the shortest contig that, together with longer contigs covers the 50% of the total genome. The NGA50 is computed similarly like NG50, but instead of considering contigs, we consider blocks correctly aligned to the reference. Both NG50 and NGA50 are presented as proportions of the maximum achievable NG50 (NGA50) values for each respective genome.











