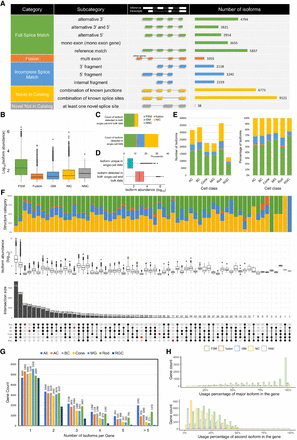
Overview of single-cell isoform catalog in the mouse retina. (A) Classification of isoforms according to their splice sites when compared to reference annotations and the number of isoforms in each category detected by the entire data set. (B) Box plot showing the isoform abundance in different categories specified in (A). The adjusted P-value shows the group differences. (C) Number and classification of isoforms detected in both single-cell and bulk RNA-seq data compared to isoforms uniquely detected using single-cell data. (D) Isoform abundance of isoforms detected in both single-cell and bulk RNA-seq data compared to isoforms uniquely detected using single-cell data. (E) Summary of number (left) and percentage (right) of isoforms in different categories for each cell class, colored by categories specified in A. (F) UpSet plot showing overlap of isoforms in cell classes, where number and percentage of isoforms shared by different cell classes were indicated in the top bar charts, colored by categories specified in (A), and the isoform abundance in each group were showed in the box plot. (G) Bar plot of the number of distinct isoforms expressed per gene. Genes with more than five distinct transcripts were merged. (H) Histogram showing the differential transcript usage in the gene of the two most abundant isoforms of each gene, grouped and colored by categories specified in A.











