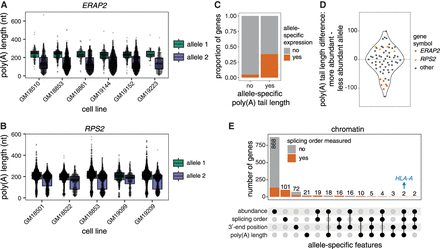
Genetic regulation of poly(A) tail length. (A,B) Examples of allele-specific poly(A) tail length in (A) ERAP2 and (B) RPS2. Each dot represents one read. A two-sided Wilcoxon rank-sum test was used to compare tail length distributions between alleles for each LCL. All tests were statistically significant (adjusted P-value < 0.001). (C) Proportion of genes that showed a skewed chromatin-associated RNA abundance ratio (allele 1 reads/total reads) out of genes with significantly different poly(A) tail lengths or not. The two groups were compared with a two-sided Fisher's exact test (P-value = 9.232 × 10−16). (D) Violin plot of the distribution of the difference in poly(A) tail length between the more abundant and the less abundant allele for genes with a skewed chromatin-associated RNA abundance ratio. Each dot represents one gene in one LCL. (E) UpSet plot showing the number of genes with skewed chromatin-associated RNA allelic abundance ratio (<0.4 or >0.6), allele-specific splicing order, significant differences in 3′-end position and/or significant differences in poly(A) tail length between alleles.











