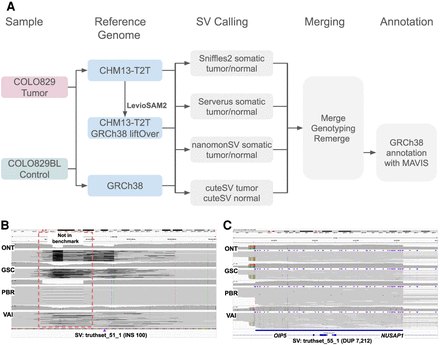
Schematic overview of the benchmark analysis and SVs examples. (A) Schematic overview of our variant calling and comparison methods using CHM13-T2T versus GRCh38. (B) IGV screenshot of an SV located in Chr 14 cataloged as missing (truthset_51_1, purple triangle) according to the benchmark by Valle-Inclan et al. We did not detect any reads supporting it. Moreover, we detected a DEL in all four samples, which is missing from the benchmark. (C) IGV screenshot of a DUP located in Chr 15 that was assigned two distinct SV types (DUP and INS), and thus cataloged as missing according to the benchmark by Valle-Inclan et al. Manual inspection in IGV showed the SV. Notice that these alignments contain mapping artifacts in the ONT samples.











