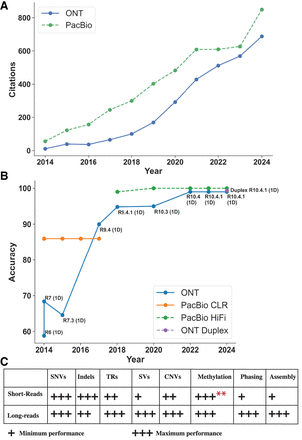
Long-read accuracy, citation trends over time, and comparison to short reads. (A) Citations of PacBio and Oxford Nanopore Technologies (ONT) long-read sequencing publications from 2014 to the present demonstrate their growing impact in the field. We collected citations from PubMed and excluded review articles. (B) This figure presents the evolution of long-read sequencing accuracy over time for ONT (Ashton et al. 2015; Goodwin et al. 2015; Laver et al. 2015; Suzuki et al. 2017; Ferguson et al. 2022; Ni et al. 2023b; Sanderson et al. 2024) and PacBio (Wenger et al. 2019; Amarasinghe et al. 2020; Logsdon et al. 2020; Oxford Nanopore Technologies 2020), illustrating their progress toward achieving >99% accuracy. For ONT, the analysis focuses exclusively on the 1D technology, with the 2014 R7 (1D) and the Duplex data points representing the median value, while the remaining points represent the mean values. We excluded ONT's 2D and 1D2 technologies because they ceased production in 2016. The plot distinguishes between PacBio's continuous long read (CLR) and high fidelity (HiFi) technologies. (C) Comparison between short reads and long reads in variant calling accuracy, methylation calling, and genome assembly (Oehler et al. 2023; Ni et al. 2023a; Dolzhenko et al. 2024; Espinosa et al. 2024; Kosugi and Terao 2024; Höps et al. 2025), where one plus represents the minimum performance and three pluses represent the maximum performance. ** Short reads required biochemical treatment and were used as the benchmark for methylation.











