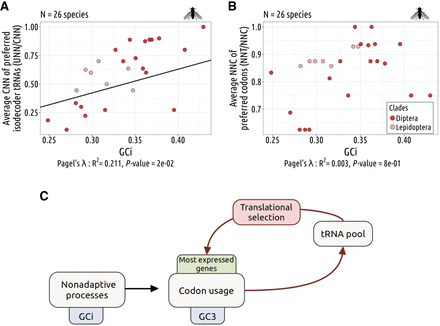
Genomic substitution patterns shape the tRNA pool. (A) Relationship between the per species CNN fraction of preferred isodecoder tRNAs (corresponding to the most abundant tRNAs) among 11 NNA/NNG synonymous codon pairs and the gene average GC in introns (GCi), for Diptera (dark red), and Lepidoptera (light red). (B) Relationship between the per species proportion of NNC preferred codons (the most overused codons in highly expressed genes compare to lowly expressed genes) among NNT/NNC synonymous codon pairs decoded by a single tRNA, along with the GCi. (A,B) Pagel's lambda model is used to take into account the phylogenetic structure of the data in a regression model (black line if significant). (C) Hypothetical schemes explaining how synonymous codon usage can be shaped conjointly by TS and by neutral substitution patterns.











