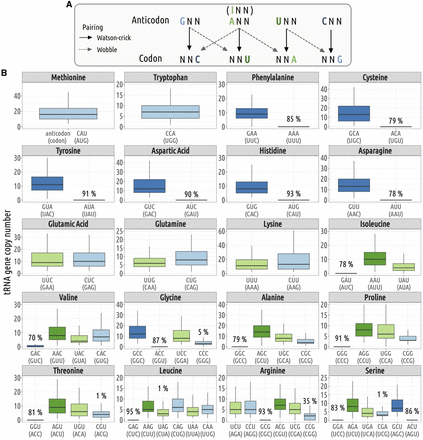
Codon–anticodon assignment. In each genome, we used the rules proposed by Percudani (2001) to assign codons to their cognate tRNA: First, isodecoder tRNAs present in a given genome are assigned to their complementary (Watson–Crick) codons, and then, the remaining codons (for which the genome does not contain any tRNA carrying the complementary anticodon) are inferred to be decoded by wobble pairing. (A) Illustration of the various possible pairings: Watson–Crick and wobble pairing. (B) Distribution of tRNA gene copy number across 223 species. The boxplot represents the median, interquartile range (box edges at 25th and 75th percentiles), and whiskers extending to the largest value no further than 1.5 times the interquartile range. For tRNA isodecoders that are absent from some genomes, the percentage of species in which they are missing is also indicated.











