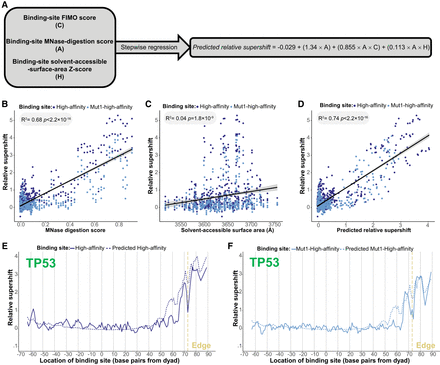
TP53 family–nucleosome binding is collectively determined by the composition, accessibility, and helical orientation of TP53 family binding sites. (A) Outline of regression model predicting TP53 family–nucleosome binding based on binding-site FIMO scores, MNase-digestion scores, and solvent-accessible-surface-area Z-scores. The resulting equation illustrates the predictive relationship between these variables and TP53 family–nucleosome binding. (B–D) Correlating the relative-supershift values (Equation 1) of TP53, TP63, and TP73 to two TP53 family binding sites with either the MNase-digestion score of these binding sites, the solvent-accessible surface area of the CATGs of these binding sites, or the relative-supershift values of TP53, TP63, and TP73 to these binding sites predicted by the multiple regression model outlined in A. (E,F) Comparison of actual versus model-predicted TP53 relative-supershift values for high-affinity and Mut1-high-affinity binding sites.











