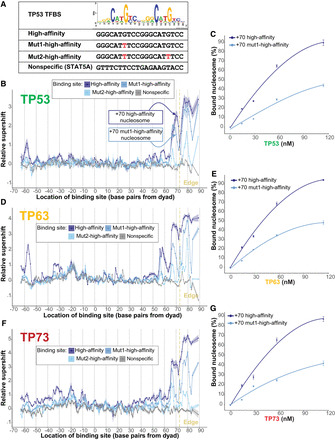
Binding site composition modulates TP53 family–nucleosome binding affinity. (A) A TP53 sequence logo above binding sites: The high-affinity site is based on the TP53 sequence logo; the Mut1-high-affinity site, on the TP53 sequence logo but with one base mutated; the Mut2-high-affinity binding site, on the TP53 sequence logo but with two bases mutated; and the nonspecific STAT5A site. (B,D,F) TP53, TP63, or TP73 binding to the four binding sites in A at different nucleosomal positions, measured relative to TP53 family binding to nonspecific binding sites and represented by relative-supershift values (Equation 1). A nonspecific motif for STAT5A is shown for comparison in gray. (Edge) The right end of the 146 bp nucleosome core particle. Shading indicates SEM. n = 3. (C,E,G) Assessing the binding affinity between TP53, TP63, or TP73 and the +70 high-affinity nucleosome or the +70 mut1-high-affinity nucleosome, both of which are indicated in B. Bound nucleosome (%) was calculated via gel-shift assays featuring Cy5-labeled nucleosomes.











