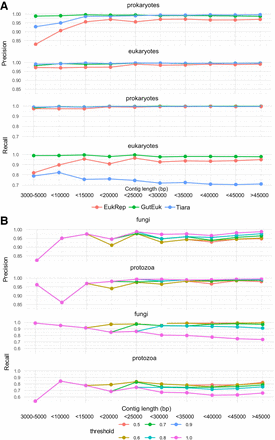
Benchmarking the performance of GutEuk, EukRep, and Tiara for contigs. (A) The performance of GutEuk, EukRep, and Tiara in differentiating prokaryotic and eukaryotic contigs of different lengths. EukRep and Tiara were used with their default settings. GutEuk was run at a pt1 of 0.5. (B) The performance of GutEuk in further differentiating fungal and protozoal contigs at different confidence levels (pt2) and contig lengths.











