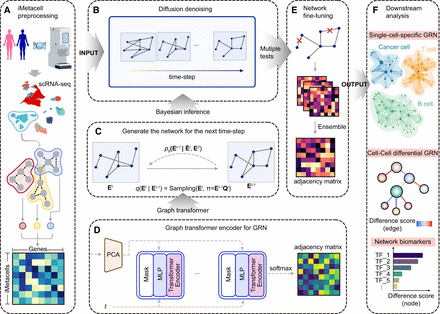
Overview of DigNet. It generates a cell-specific gene regulatory network (GRN) and extracts differential network structures from single-cell gene expression profiles. (A) Data preprocessing of human tissue scRNA-seq data. (B) Network diffusion denoising across time-steps in DigNet. (C) Transforming the adjacency matrix exported by the encoder through Bayesian inference to predict the GRN at the subsequent time-step. (D) Utilizing a transformer to encode GRN for scRNA-seq data based on the current time-step information. The procedures in C and D are repeated with each time-step to progressively denoise the network structure. (E) Correcting the network linkages (removing incorrect regulatory interactions) and integrating multiple diffusion-generated networks to produce the final cell-specific network. (F) Once the generated network structure is obtained from scRNA-seq data using DigNet, multiple downstream analytics can be performed to identify key network features driving cell heterogeneity or biomarker signatures indicative of cancerous states.











