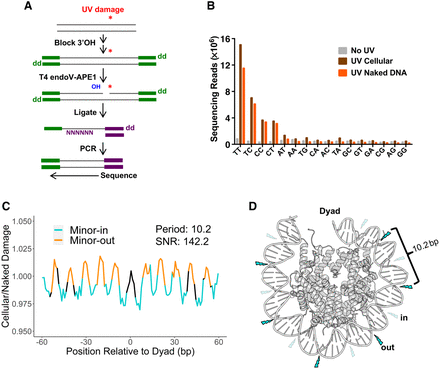
CPD-seq reads are enriched at dipyrimidines and minor-out positions in Drosophila. (A) An outline of the CPD-seq protocol used to generate a map of UV-induced CPDs in Drosophila. (B) Counts of CPD-seq reads from Drosophila S2 cells (with and without UV exposure) and UV-irradiated naked DNA, stratified by the dinucleotide sequence at the putative CPD positions. (C) Damage enrichment with respect to rotational positioning within nucleosomes. Data are normalized by a UV-exposed, naked DNA control. (D) Model of how rotational positioning impacts CPD formation in Drosophila nucleosomes. The nucleosome structure corresponds to Protein Data Bank (PDB; https://www.rcsb.org) entry 2PYO (Clapier et al. 2008). Minor-in positions are labeled with transparent lightning bolts to represent low rates of CPD formation, and minor-out positions are labeled with opaque lightning bolts to represent high rates of CPD formation.











