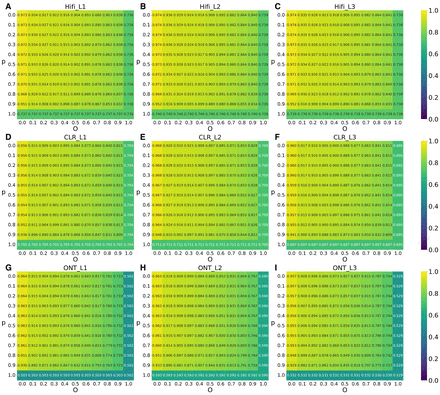
Deletion F1 accuracy of FocalSV(auto) by tuning different evaluation parameters (p and O). O is the minimum reciprocal overlap between SV call and gold standard SV. p is the minimum percentage of allele sequence similarity between SV call and gold standard SV. O and p vary from 0 to 1 with a 0.1 interval. (A–C) The F1 heat map for deletions by FocalSV(auto) on three HiFi data sets. Every cell in the heat map represents the F1 score under a specific pair of p and O evaluation. (D–F) The F1 heat map for deletions by FocalSV(auto) on three CLR data sets. (G–I) The F1 heat map for deletions by FocalSV(auto) on three ONT data sets.











