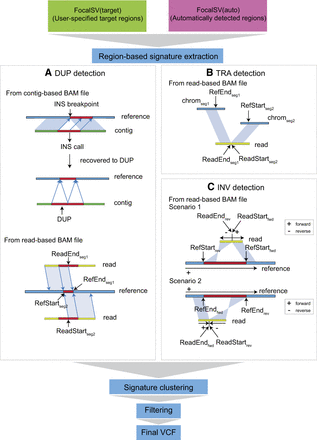
Schematic diagram of duplication, translocation, and inversion breakpoint signatures detected by FocalSV. (A) Duplication: duplications can be identified using both contig-based and read-based BAM files. In a contig-based BAM file, a duplication is detected when an insertion call shows the alternate allele mapped to the surrounding sequence of the insertion breakpoint. In a read-based BAM file, a duplication signature is identified when two adjacent segments of a read align to overlapping regions on the reference genome in the same orientation. (B) Translocation: translocations are inferred when two adjacent segments of a read align to different chromosomes. (C) Inversion: inversions are detected when two adjacent segments of a read align in opposite orientations. Additional details on detecting duplications, translocations, and inversions can be found in the Methods section.











