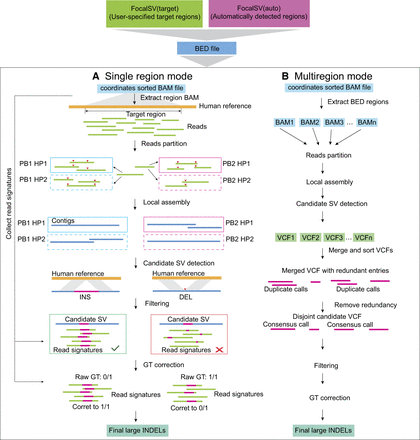
Schematic diagram of the FocalSV large indel detection pipeline. The workflow for large indel detection includes two modes: single region mode, and multiregion mode. (A) Single region mode: the input data include a high-quality reference genome and a BAM file containing aligned long reads. The reads extraction module isolates long reads aligned to the region of interest. The haplotyping module partitions these reads into distinct parental haplotypes. The local assembly module uses the phased reads to perform independent de novo local assemblies. Finally, the variant calling module identifies indel structural variants (SVs) by comparing the assembled contigs to the reference genome, followed by filtering and genotype (GT) correction in postprocessing steps. (B) Multiregion mode: the input data includes a high-quality reference genome and a BAM file containing aligned long reads. FocalSV retrieves region-specific BAM files and processes each region independently through reads partitioning, local assembly, and SV detection. The VCF files from all regions are merged into a single file, and redundant variants are removed using a clustering algorithm. SV filtering and genotype refinement are then applied to produce the final VCF file.











