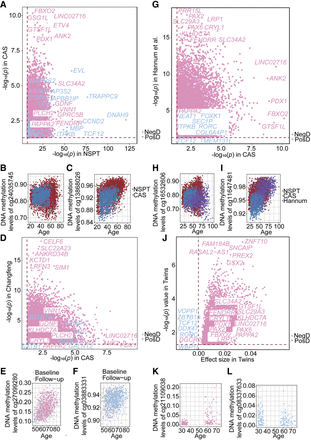
Replication of drift-CpGs in independent cohorts. (A) Replication of significant drift-CpGs in CAS, with thresholds corresponding to Bonferroni significance in NSPT and nominal significance in CAS. CpGs are colored by drift direction and selected genes are labeled. (B,C) Scatter plots of top positive (B: cg24035745) and negative (C: cg13868026) drift-CpGs validated in CAS. (D–F) Validation of drift-CpGs in the Changfeng cohort, including representative positive (E: cg27099280) and negative (F: cg03883331) examples. (G–I) Replication in Hannum et al. (2013), with representative positive (H: cg16532606) and negative (I: cg11647481) drift-CpGs. (J–L) Validation in monozygotic Danish twins (Tan et al. 2014), including top positive (K: cg21109038) and negative (L: cg08337633) drift-CpGs.











